Bridging the gap between High Performance Computing and Big Data

In today’s blog post, I’ll talk about the successful (yaaaay!) progress of my project with a technical point of view. So if you are into Computer Science, I’m sure you will find this interesting! This post will be structured using some small pieces of the scientific article we’re writing, but of course I can’t write everything about the research here.
Introduction
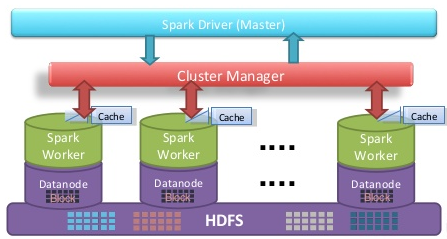
Figure 1: Apache Spark architecture
Apache Spark applications written in Scala run on top of the JVM (Java Virtual Machine) and because of this they cannot match the FPO performance of Fortran/C(++) MPI programs which are compiled to machine code. Despite this, it has many desirable features of (distributed) parallel applications such as fault-tolerance, node-aware distributed storage, caching or automated memory management (see Figure 1 for an overview of the Apache Spark architecture). Yet we are curious about the limits of the performance of Apache Spark applications in High Performance Computing problems. By writing referential code in C++ with MPI and also with GPI-2 for one-sided communication, we aim to carry out performance comparisons.
We do not expect the resulting code to be, in terms of performance, truly competitive with MPI in production applications. Still, such experiments may be valuable for engineers and programmers from the Big Data world who implement, often computationally demanding algorithms, such as Machine Learning or Clustering algorithms.
Some literature review
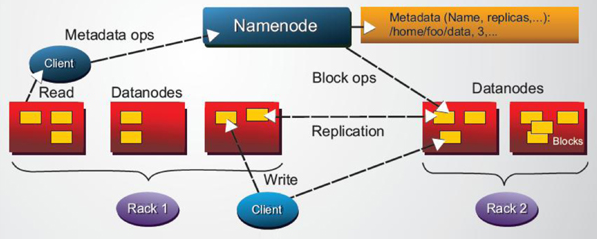
Figure 2: HDFS architecture
In the same way as was mentioned in previous work, it has been widely known that High Performance Computing frameworks based on MPI outrun Apache Spark or HDFS-based (see Figure 2 for an overview of the HDFS architecture) Big Data frameworks, by usually more than an order of magnitude for a variety of different application domains, e.g., SVMs & K-nearest neighbors clustering, K-means clustering, graph analytics, and large scale matrix factorizations. Recent performance analysis of Spark have shown that computing load was the main bottleneck in a wide number of Spark applications, particularly during the serialization and deserialization time.
It has been shown that it is possible to extend pure MPI-based applications to be elastic in the number of nodes using periodic data redistribution among required MPI ranks. However, this assumes that we are still using MPI as the programming framework, hence we do not get the significant benefits of processing with Apache Spark required for large scale data, such as node fault tolerance.
Unlike previous approaches described in previous work, in what we are investigating we propose a fair comparison, with similar implementations of algorithms in Spark on HDFS and in C++ on MPI. We will run both algorithms with distributed data along commodity cluster architectures inside a distributed file system, using a one-sided communication approach with native C libraries that create an interface to encapsulate the file systems functionalities so as to preserve node fault tolerance characteristics exhibited by Spark with HDFS implementations.
Partitioned Global Address Space
To provide our non-Spark applications with a Fault Tolerance features, we’ve used the GASPI API for the Partitioned Global Address Space (PGAS).

Figure 3: GASPI one-sided communication segment-based architecture
GASPI is a communication library for C/C++ and Fortran. It is based on a PGAS style communication model where each process owns a partition of a globally accessible memory space. PGAS (Partitioned Global Address Space) programming models have been discussed as an alternative to MPI for some time. The PGAS approach offers the developer an abstract shared address space which simplifies the programming task and at the same time facilitates data-locality, thread-based programming and asynchronous communication. The goal of the GASPI project is to develop a suitable programming tool for the wider HPC-Community by defining a standard with a reliable basis for future developments through the PGAS-API of Fraunhofer ITWM. Furthermore, an implementation of the standard as a highly portable open source library will be available. The standard will also define interfaces for performance analysis, for which tools will be developed in the project. The evaluation of the libraries is done via the parallel re-implementation of industrial applications up to and including production status.
One of the proposed benchmarked algorithms: K-Means clustering
To benchmark the different approaches we propose, we have implemented some algorithms, one of them being K-Means clustering. K-Means clustering is a technique commonly used in machine learning to organize observations into k sets, or clusters, which are representative of the set of observations at large. Observations (S) are represented as n-dimensional vectors, and the output of the algorithm is a set of k n-dimensional cluster centers (not necessarily elements of the original data set) that characterize the observations. Cluster centers are chosen to minimize the within-cluster sum of squares, or the sum of the distance squared to each observation in the cluster:
![]()
where Si is the set of observations in the cluster i and ui is the mean of observations in Si.
This problem is NP-hard and can be exactly solved with complexity O(ndk+1 log n). In practice, approximation algorithms are commonly used to get results that are accurate to within a given threshold by terminating before finally converging, but these algorithms can still take a significant amount of time for large datasets when many clusters are required.
The main steps of K- means algorithm are as follows:
- Select an initial partition with K clusters; repeat steps 2 and 3 until cluster membership stabilizes.
- Generate a new partition by assigning each pattern to its closest cluster center.
- Compute new cluster centers.
The following illustration shows the results of the K-Means algorithm on a 2-dimensional dataset with three clusters:
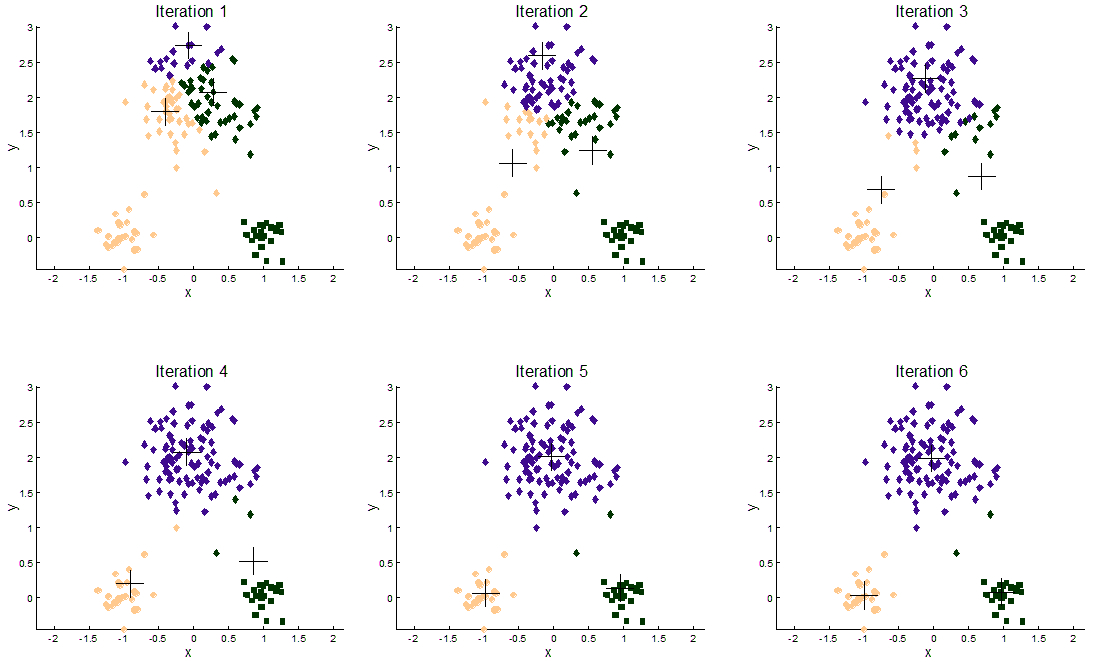
We’ve tested our implementations of the algorithm using different datasets, some artificial and some real data obtained from some bioinformatics studies from researchers at the Biology Faculty of the Slovak Academy of Sciences. The artificial data goes up to more than 10 million samples with 2 dimensions for some instances (to benchmark computational time), and also some high dimensional (more than 100 dimensions) instances (to benchmark time resilience).
So that’s all for today, if you are really interested about the work that’s going on, just wait for the next blog post or for the article to be published (if we’re lucky enough). Here is a video of the city of Bratislava celebrating my progress with some fireworks:

[…] successfully implemented – as my previous post explains, the proposed algorithms with different traditional HPC approaches (MPI) and with renowned […]