HPC Implementation of Molecular Surfaces
Project reference: 2132
Molecular surfaces have tremendous predictive power in terms of evaluating the interaction with potential binding partners, e.g. small molecules, drugs, biochemical ligands, antibodies, signaling molecules and many more. Consequently, an efficient method to produce such molecular surfaces in a robust yet general way will find many applications in contemporary science ranging from computational biology (PB, MM/PBSA, cryo-EM, Struct. Bio) to physics (Maxwell’s Eq., CFD, MD).
The current project aims to continue the efforts already inititated in last years SoHPC project 2024, where a novel approach based on Marching Tetrahedra had been developed from scratch. Particularly encouraging were the edge-based formulation, the principal availability of an efficient method to identify closed subsets of surfaces, the analytical classification using the Euler characteristics and first ports to the GPU. Given all these remarkable achievements (made by even more remarkable SoHPC students), it is only natural to pursue an extension of these activities also in this years edition of the SoHPC 2021 program. Ideally, one would take over as much as possible from the previous implementation. However, this is not a binding constraint and depending on the outcome of initial validations every aspect of the current approach may become subject to change. Anticipated ameliorations are thought in the following areas: (i) general applicability to any molecular structure defined in pdb format, (ii) enhanced robustness, (iii) proven smoothness of the molecular surface devoid of any internal artefacts (iv) significantly scaled-up size coverage. Target platform will again be the GPU, as it holds strongest promise in delivering the required performance to cope with large-scale biomolecular structures.
The proposed activitiy includes a great variety of individual tasks (which can be handed out independently in accordance with trainees’ skill level and interest) starting from simple geometric considerations and encompassing all essential steps of the development cycle characteristic of contemporary scientific software development.

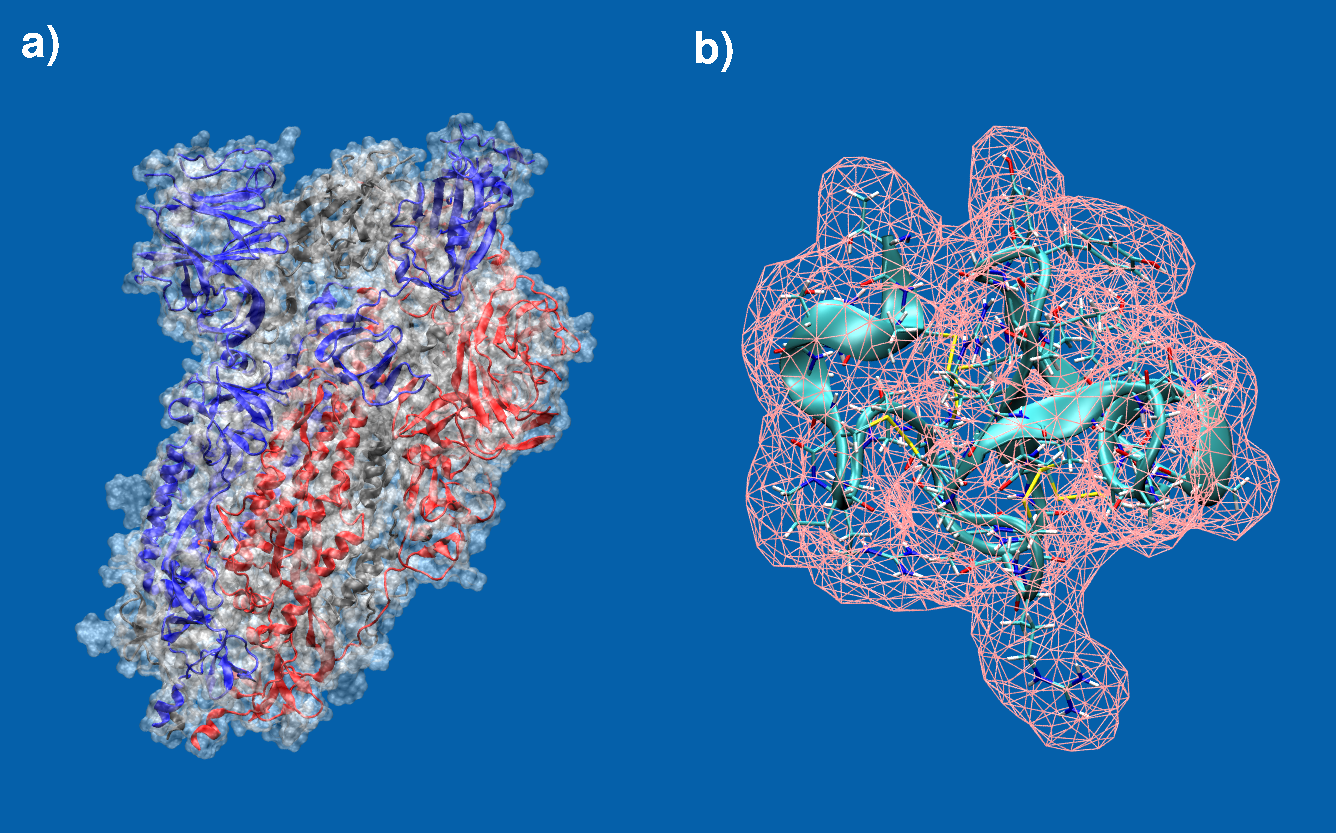
Molecular surfaces of a) the spike protein in SARS-CoV-2 and b) EAFP2, an antifungal peptide. Students from last years SoHPC project 2024 have already implemented a fully functional molecular surface program for which results are shown in panel b). However, this approach needs to further be validated, made more robust, re-examined and fine-tuned and scaled up considerably to also cover large sized biomolecules such as for example the one shown in panel a) which is approximately 45x the size of b).
Project Mentor: Siegfried Hoefinger
Project Co-mentor: Markus Hickel and Balazs Lengyel and David Fischak
Site Co-ordinator: Claudia Blaas-Schenner
Participants: Miriam Beddig, Ulaş Mezin
Learning Outcomes:
Familiarity with basic development strategies in HPC environments. A broader understanding of fundamental key algorithms in scientific computing and how to develop corresponding implementations in an efficient way.
Student Prerequisites (compulsory):
Just a positive attitude towards HPC for scientific applications and the readiness for critical and analytical thinking.
Student Prerequisites (desirable):
Familiarity with Linux, basic programming skills in C/C++/Fortran, experience with GPUs, basic understanding of formal methods and their translation into scientific applications;
Training Materials:
Public domain materials and some web information about CUDA.
Workplan:
- Week 1: Basic HPC training; explore local HPC system;
- Week 2: Theory and first runs with previous code;
- Week 3: Workplan formulation;
- Weeks 4-7: Actual validation, refinement, fine-tuning, performance assessment and upscaling;
- Week 8: Write up a final report and submit it;
Final Product Description:
Ideally we get a robust and super-efficient molecular surface program implemented on the GPU that can address biomolecules like the spike protein of SARS-CoV-2 in less than 0.02 sec, but even more important, are summer students having gained good experience with practical work in HPC environments.
Adapting the Project: Increasing the Difficulty:
Increasing performance gains in absolute terms as well as relative to existing implementations;
Adapting the Project: Decreasing the Difficulty:
Various optional subtasks can either be dropped or carried out in greater detail.
Resources:
Basic access to the local HPC infrastructure (including various GPU architectures) will be granted. The codebase from last years SoHPC project 2024 is readily available.
Organisation:
VSC Research Center, TU Wien


Leave a Reply