Time to say Goodbye after Optimizing

After 2 months, I came to the end of Summer Of HPC. I would like to share with you the optimization methods I followed in this two-month great adventure. In my previous blog post, I explained how the function compute_density_at_grid_points() works. Before I clarify the optimization methods, I strongly recommend that you take a look at my previous blog post so that you can better understand the effects of optimization metrics.
OPTIMIZATIONS
I would like to share the optimization I applied with you.
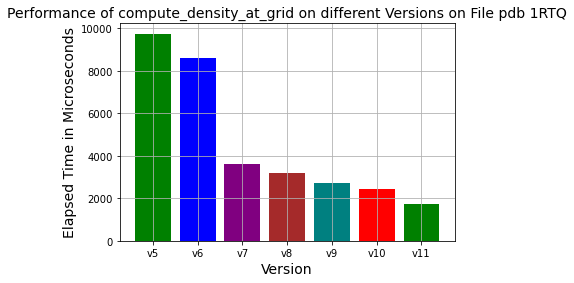
Here version 5 refers to last years implementation. All individual optimization steps showed continuous progress towards improved runtime conditions.
Version 6
In this version, the device function pointer is used to eliminate if statements. The achieved performance improvement was on the order of 10% (see the chart, blue versus the green bar).
Version 7
Slater densities are calculated according to different atom types. While calculating densities, different constant variables are created according to orbitals and atomic numbers every time the density functions are called. These are pre-calculated and assigned fixed values instead of being re-calculated in the density functions’ calls. Acceleration was observed at approximately 300%(see the purple bar in the chart).
Version 8
Density calculations include as many exponential operations as the number of shells of the atom. In addition, after calculating the distance at each grid point, a power operation is performed. These processes are very costly compared to the Fast-Math library. For this reason, the Fast-Math library is used. In addition to the seventh version, an acceleration of about 33% was achieved(see the dark-red bar in the chart).
Version 9
In the profiling results, the number of instructions for creating and deleting local variables in each device function consists of most of the instructions compared to the total number of instructions. For this reason, by following the method of minimizing function calls, all functions were gathered under a single function, namely, compute density at grid points. Acceleration was observed approximately 50% in addition to the eighth version(compare cyan to the dark-red bar in the chart).
Version 10
If statements were revised from the most frequent atom to the least frequent atom. It provides us with an additional 20% performance speedup compared to the ninth version(see the red bar in the chart).
Version 11
Data types were converted from double to float since 7 decimal points are sufficient for numerical accuracy. It provides us with an additional 20% performance speedup compared to the tenth version(see the dark-green bar in the chart).
The obtained results show between 8x acceleration for larger sized molecules and 13x acceleration for smaller sized molecular structures.
The End
During these two months, I improved my skills by addressing different topics with different approaches every day. I had a very enjoyable 2 months. I am very happy to participate in the Summer of HPC 2021. Thank you to everyone who gave me this opportunity.

Leave a Reply