Transformers Model for Multi-Label Text Classification!
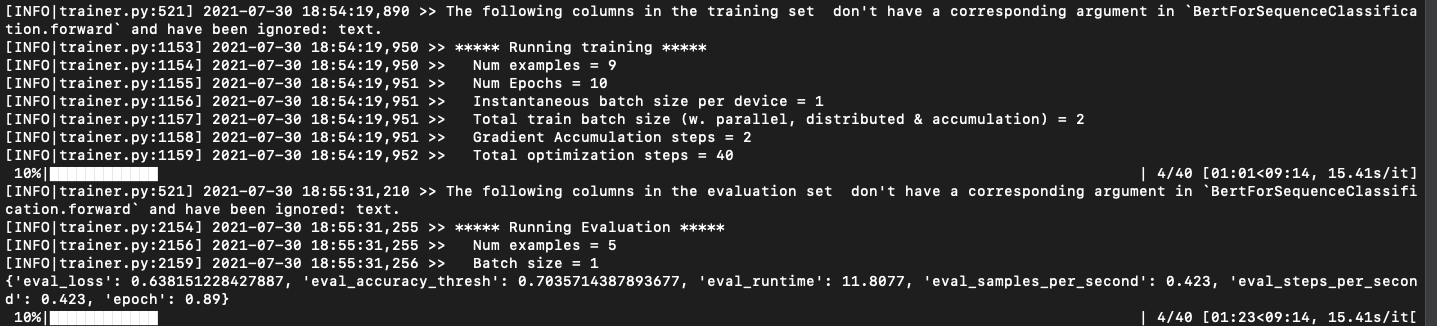
Hello again! As I mentioned in my previous blog and week3 report, I work on a multi-label classification model on biomedical texts, during a couple of weeks I worked on how to fine-tune transformers models (https://huggingface.co/models) for required tasks.
First of all, it was really tough to train the initial model for the first time than it seems. It requires you to go dive into model architectures, understand parameters, what is going on behind the scenes, and, more importantly, implement it. I will try to explain the steps I followed and what we have achieved so far and also will try to give some institutions and resources if you are interested in more about what transformer models are.
To give a quick introduction to transformers models:
Transformer models are used to solve all kinds of NLP tasks, like the ones mentioned in the previous section. Here are some of the companies and organizations using Hugging Face and Transformer models, who also contribute back to the community by sharing their models:
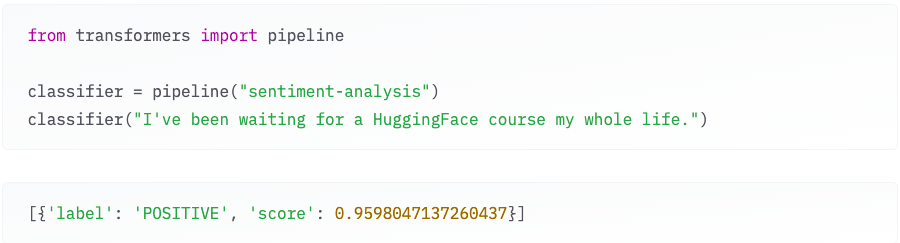
Figure 1: An example of out-of-box one of the transformers models for sentiment-analysis
Step 1:
Before I talk about the first step, I want to give a brief introduction to what HuggingFace(https://huggingface.co/) is for and why we use it. As the first step, we needed to upload our dataset into the HuggingFace datasets environment (https://huggingface.co/datasets). Even if you don’t need to upload any datasets, it is sort of a convention for using HuggingFace environment like if you don’t want to spend a lot of time on uploading or dataset structure, I strongly recommend you to upload your dataset.
Our dataset includes text and label features and actually, that is all that we need to be able to train pre-trained language models for the multi-label classification tasks.
One of the inputs in the training dataset:
{"articles":[
{ "journal": "Mundo saúde (Impr.)", "title": "Determinantes sociales de la salud autorreportada: Colombia después de una década", "db": "LILACS", "id": "biblio-1000070", "decsCodes": [ "21034", "13344", "3137", "50231", "55453" ], "year": 2019, "abstractText": "El objetivo de este estudio fue analizar los determinantes sociales que afectan el estado de salud autorreportado por la población adulta colombiana. Con los datos de la Encuesta de Calidad de Vida realizada por el Departamento Administrativo Nacional de Estadísticas, DANE, en el año 2015, se estimaron modelos de respuesta cualitativa ordenada para evaluar el efecto de un conjunto de determinantes sociales sobre el estado de salud. Los resultados señalan que los factores que afectan positivamente el autorreporte de buena salud son el ingreso mensual del hogar, los incrementos en el nivel educativo, ser hombre, habitar en el área urbana y en regiones de mayor desarrollo económico y social. Los factores que incrementan a probabilidad de autorreportar regular o mala salud son el incremento en la edad, no tener educación, ser mujer, ser afrodescendiente, estar desempleado, vivir en la región pacífica y en la zona rural. Más de una década después de los primeros estudios sobre los determinantes de la salud realizados en el país, los resultados no evidencian grandes cambios. Las mujeres, los afrodescendientes y los habitantes do Pacífico Colombiano son los grupos de población que presentan mayor probabilidad de autorrelatar resultados de salud menos favorables."}
]
}Step 2:
Before the tokenization process for the “text” feature, we need to adjust dimensions to be able to feed to the model or give the right formatted input that what the model expects. To do this we need to define how many unique labels we have among all training, validation, and test datasets. After defining this we needed to convert label feature into binary unique labels length format such as the above example that has [ “21034”, “13344”, “3137”, “50231”, “55453” ] decsCodes which also can be called class or label.
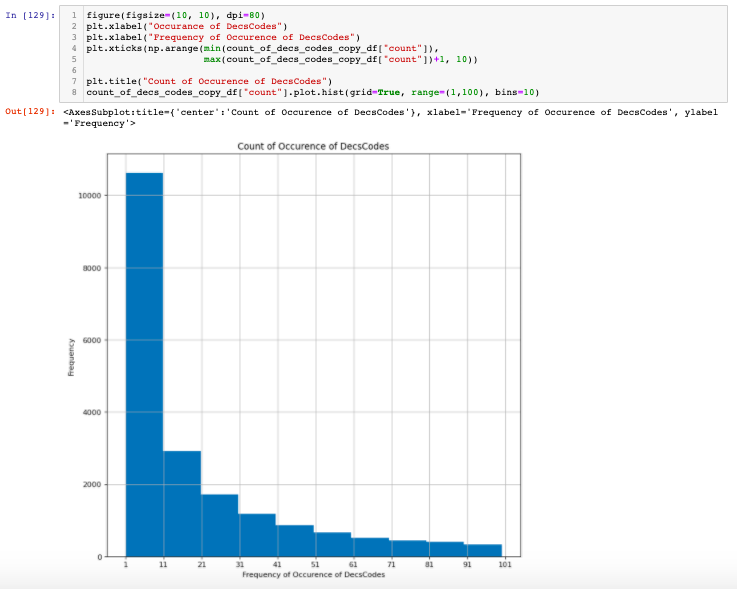
Figure 2: Frequency of DeCS Codes.
As we can see that many of the DeCS Codes have a very low count. For the scope of this problem, we could restrict ourselves to less than the amount of unique DeCS Codes. That gives us still the same 370 thousand rows of medical texts which are decent enough given that we are using pre-trained models.
The histogram plot reveals that there are almost 10 thousand unique DeCS codes that occurred in less than 10 in the entire dataset. Also in general, reducing that many unique codes still could be reasonable for the model to be able to perform classification.
After defining this we needed to convert label feature into binary unique labels length format such as the above example that has [ “21034”, “13344”, “3137”, “50231”, “55453” ] decsCodes which also can be called class or label.
[ “21034”, “13344”, “3137”, “50231”, “55453” ] ==> [1,1,1,1,1,0,0,0,0…]1xlength_of_labels

Step 3:
After some tokenization process, we need to train our model based on tokenized text and label inputs!
Step 4:
After training step, we need to evaluate our performance based on the test set that is already labeled and maybe change some hyperparemeters and tweak the model for better accuracy,
Model Name | Epoch | Accuracy |
mBERT | 1 | 0.873 |
| BETO | 1 | 0.8723 |
BioRoBERTa | 1 | 0.8704 |
Table 1: Accuracy Results Table for Epoch 1
| Model Name | Epoch | Accuracy |
mBERT | 4 | 0.8596 |
| BETO | 4 | 0.8613 |
BioRoBERTa | 4 | 0.8716 |
Table 2: Accuracy Results Table for Epoch 4
According to the final results, each model have very close accuracy to each other. Increasing epoch from 1 to 4 does not improve accuracy much as the models might have started to overfit the training dataset more thus accuracy might be started to reduce at some point. To overcome this obstacle, EarlyStoppingCall- back class from the Huggingface library can be used for future improvements to be able to prevent and stop learning once models might reduce the accuracy. If you have any questions I am more than happy to answer! You can find a more detailed explanation on my final report, thank you for your attention! 🙂
This was such an entertaining and informative summer for me, I would like to thank my mentors for making this program very insightful! I strongly recommend the Summer of HPC program to people who are interested in HPC!

Leave a Reply