First week in Copenhagen

First Meeting and Introduction
When Moti and I met in front of the main building of Niels Bohr Institute (NBI), it was no more than 17 hours since we arrived to Copenhagen. A joint breakfast was waiting for us together with Mads, our project mentor, and Brian, NBI site coordinator, inside Brian’s office (PS we didn’t receive any puppies or gifts…).

the main building of Niels Bohr Institute
Following the breakfast, Mads presented a basic physical and algorithmic background of the project, pointing to the high complexity and HPC needs.
The project itself is split into two main parts:
-
Develop a k-means clustering algorithm for 6-dimensional space.
-
Parallelize the k-means clustering implementation to utilize all HPC capabilities, such as multi-core, vectorization and MPI processes.
Soon after the ground was set, we were handed a paper, a Bachelor Thesis, prepared by a local student and summarizing previous work on this topic, including GPGPU implementations with CUDA/OpenCL. The paper gave us a good feeling of the challenge and how complex the task could be.
Both of us will be collaborating together during the summer on the two projects. To improve our productivity some time was spent on setting a shared workspace to exchange code and version control (important!). It was accomplished via Git (distributed version control and source code management).
To end up on a good note, our first day was finished with a dinner, including most of the members of the eScience group, here at Copenhagen. That was a good time (We can recommend some local beers to future visitors).
What k-means clustering is and why we want to use it?
We want to do some physical computations with particles in a space, but we have too many particles in the space, so many that it is impossible to compute. It is a problem to store particles in memory, so we want assigned particles which are close to one bigger particle. Let’s call that bigger particle a cluster. The cluster will represent all group of individual particles.
How to find these clusters? K-means clustering! K-means clustering is algorithm which returns clustered dataset into k clusters in which each particle belongs to the cluster with nearest mean. How it works? Imagine that we have a set of particles, for simplicity just in 2-dimension like you can see at the image below (left). Let’s say that we want to have 4 clusters. Each cluster is represented by its center and particles which belongs to the cluster. In the beginning we can put centers of clusters randomly into space. Then we assign the nearest cluster to each particle, after that we shift center of each cluster to its mean. These two steps, assignment and relocation, we repeat until the cluster no longer change. See the result (right).
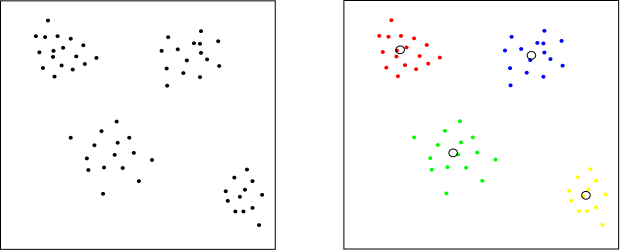
Dataset clustered to 4 clusters.
Study of K-Means Algorithms
Although the first task of the project was to develop a k-means algorithm, it seemed like a tedious task at the beginning. Therefore, the next couple of days were dedicated for studying this subject into more detail, evaluating different algorithms and approaches to the problem of clustering.
We ended up with a collection of nearly 22 papers describing core characteristics of different implementations and variations of the k-means algorithm. In addition to the basic Lloyd’s implementation there are others which perform faster under some circumstances or provide algorithmic optimization.
Programming & Development of K-Means
Luckily, we were able to find implementations for most of the proposed algorithms & variants. Our main project for developing a k-means algorithm was set to integrate these libraries.
Need to be mentioned, the development environment is Linux (Mint for Lukas, Fedora for Moti), Eclipse as IDE (with C/C++ support) and the compiler is simply g++ (4.7.2) for now.
All integrates with Git.
The visualization software we employ is octave (or Matlab under Windows). Compared with other sophisticated tools, it provides us simple means to plot 2D figures without the burden of complex systems.
The first two libraries to integrate were kmlocal (http://www.cs.umd.edu/~mount/Projects/KMeans/, Developed by Prof. David Mount, Computer Science, University of Maryland, http://www.cs.umd.edu/~mount/) and KMeans++ (http://code.google.com/p/p2pdmt/). Both are serial libraries for k-means intended for academic research. Kmlocal contains 4 variants: basic Lloyd’s, Swap, Hybrid and Easy Hybrid. KMeans++ contains naïve k-means and k-means++ algorithms.
Those libraries are written primarily in C++, however, the main scientific application is written in FORTRAN and some of the previous work was done in Python. We decided to continue with C++, which both of are familiar with, also considering future consequences: it is easier to integrate C/C++ with FORTRAN and most of the optimization techniques for HPC are available for C/C++ mostly (manual vectorization, OpenMP and MPI).
Despite some minor issues with pointers everything is going quite well. By the fourth day of project start, we had two libraries integrated, running and returning results for simple 2D cases:
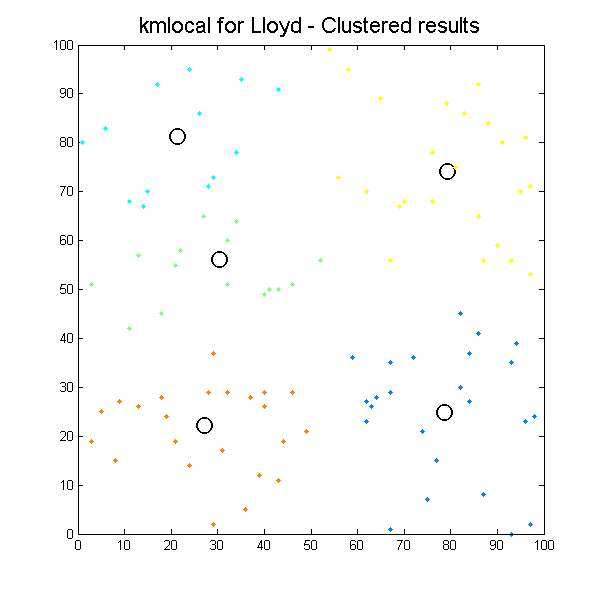 Clustered results for kmlocal_lloyd algorithm. |
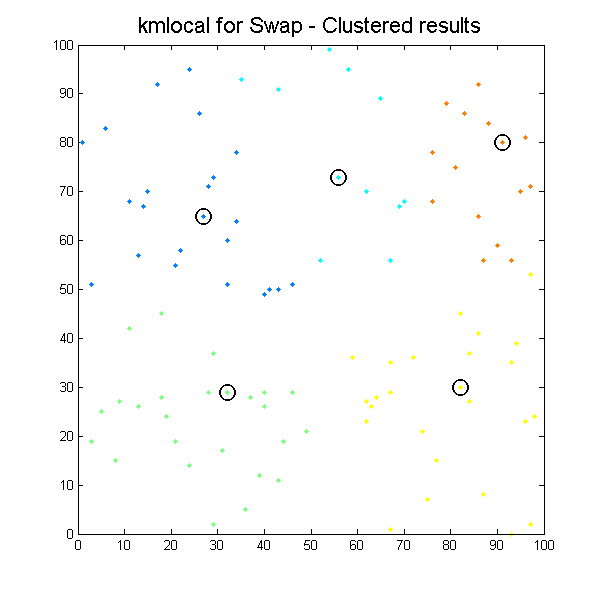 Clustered results for kmlocal_swap algorithm. |
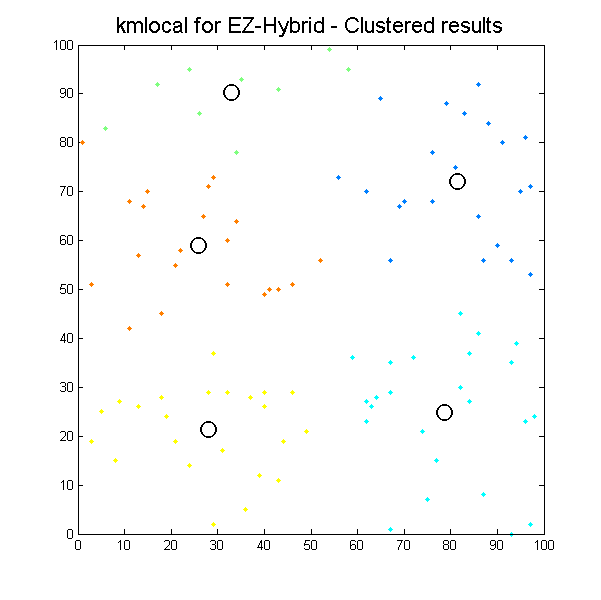 Clustered results for kmlocal_ez_hybrid algorithm. |
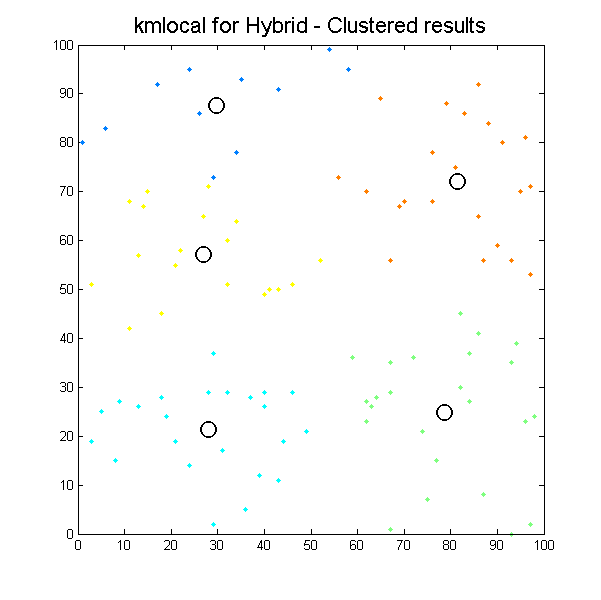 Clustered results for kmlocal_hybrid algorithm. |
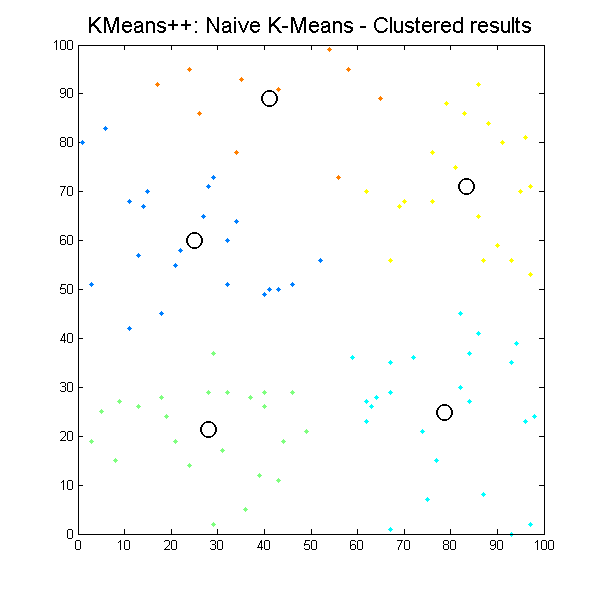 Clustered results for kmpp_naive algorithm. |
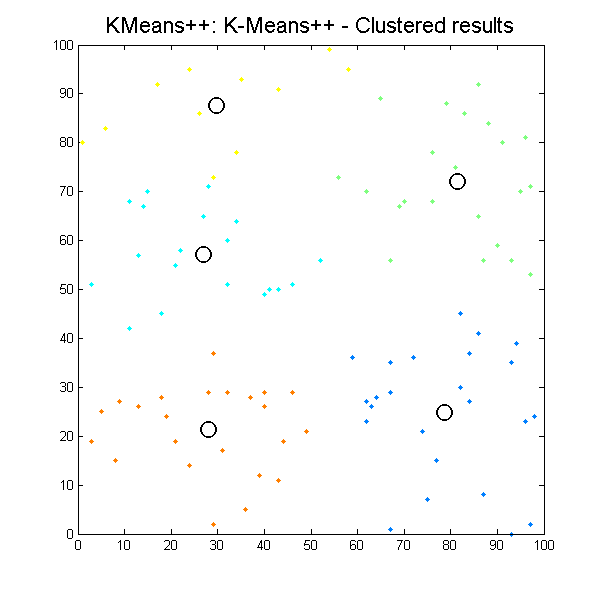 Clustered results for kmpp_pp algorithm. |
It can be seen how the results which k-means algorithms produce depend significantly on initial conditions, so we can easily get different results with the same input data. It is the issue for our next work, define initial conditions to get the most suitable results as possible.
Future Work
Our plans for the upcoming week are mostly to move forward with implementation and provide more accurate performance profiling of the algorithms. For that, results of each library has to be physically validated (as of ambiguity problems). On another hand, each algorithm will be profiled independently for comparison and future runtime performance optimizations.
As a first measure, we need to run the algorithms on actual, physically realistic data. Clustering problems are very domain sensitive, therefore it is usually meaningless to compare algorithm results based on random or non-relevant data as results might be false and hard to interpret (in fact, interpretation is flavor dependent, of specific need).
Using a realistic dataset we could also benchmark the algorithms for runtime performance (computation time, bottlenecks, memory usage and complexity), as different implementations pose varying complexity based on the different parameters.
For this task, the main cluster will be used to create a gold reference, both for quality of results, and a comparative performance baseline.
Be aware that current algorithms were compiled using g++ (4.7.2), obviously performance may depend heavily on compiler selection, so a small task will be dedicated to investigate after alternative compilers and compare results (such as Intel(R) compiler or PGI).
So, stay tuned for more algorithms, benchmarks and comparisons in the upcoming week!

[…] and I work on the k-means clustering project, you can read our first post about it. Now we prepared a short and easy quizz related with k-means topic for […]