Journey in the SoHPC’21 and Genomicper Package
Hi everyone! I hope all you are being-well. Before I tell my SoHPC adventure with you, I would like to briefly inform you about our project which is called “Re-engineering and optimizing software for the discovery of gene sets related to disease”.
Analysing and interpreting biological data in a computer environment have been made it possible by help of the developing technologies. This technology is used in kinds of different and wide areas, from network and system biology to drug researchs. In this project the affect of genetic factors with pathway analysis on the disease susceptibility on the genetic basis level has been investigated. The motivation bedin the project is optimizing the Genomicper package, which was written was previously written in R programming language, to enable it to work with larger data sets. Because as the data size increases, the number of genes that can be examined and therefore the diseases that can be detected also increases. Besides, resent technologies provides an opportinity to work with larger data sets.
The Genomicper Package Optimisation
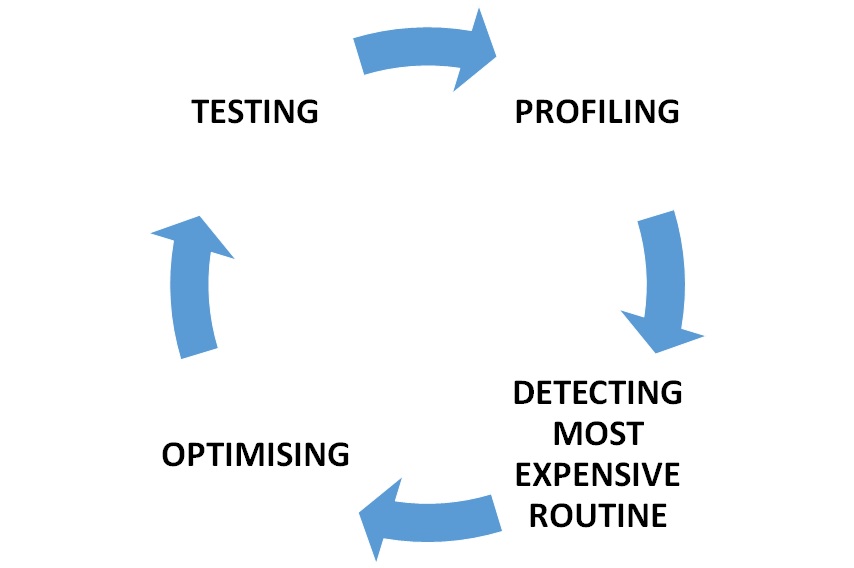
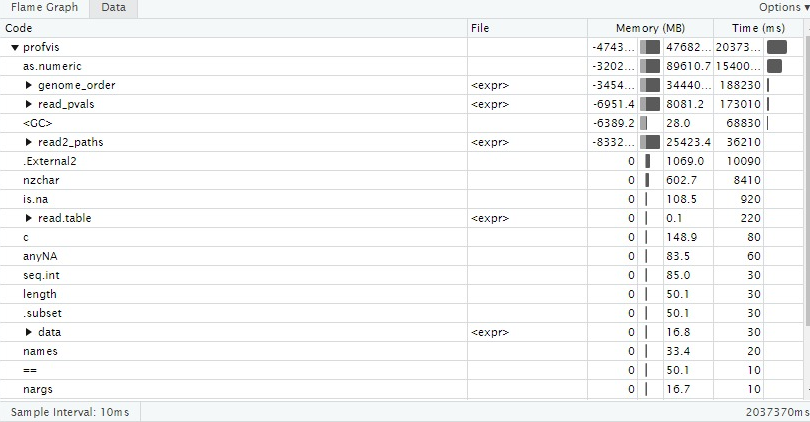
Optimization process starsts by profiling the code to detect most expensive routine in the Genomicper package according to the process time and memory usage. Then the code is optimized by algorithmic or with aim of making package use less memory if possible and to run faster. After these change, the outputs of the code will be checkedto be correct. The rewritten code is tested with different sizes of the data sets to detect the next block to be optimized.
Future Plan
Optimization process above will be done to until no more meaningful improvements can be made to the code. Then the Genomicper package will be parallized in R language.
See you next blog posts!

Leave a Reply