Parallel Flow-Cutter performances and conclusion

Hello there, I hope you’re ready to enter into the practical implications of our theoretical algorithm we saw last time. To refresh your mind, we want to find an elimination order of a graph that leads to lowest tree-width. To do so, our program will search a node separator using the flow-cutter algorithm with random inputs then cut the graph in several parts and repeat the process on sub-graphs until they are complete or tree. In practice, the flow-cutter algorithm is called multiple time, with random inputs, to extract the best possible separator and not just once which is too random. In order to manage the number of calls to the core flow-cutter algorithm, we use a parameter named nsample .Its default value is set to 20. Those calls are independent if we manage to work on several copies of our graph instead of references only. In this case, we would face some side effect with vertices and edges created and removed. Therefore, we can completely parallelize this section of code to gain computation time. To implement parallelism in Julia, we have several options but according to my lectures, there are mainly two of them:
- Use threads with the Threads.@threads macro to parallelize the loop containing the sampling of input, the call to flow-cutter , etc.
- Use coroutines/tasks with asyncmap-like functions to parallelize the calls to flow-cutter with known inputs
I’ve done some tests to compare both methods. I’ve run the two versions of the whole algorithm 20 times on the Sycamore 53 20 graph and I measured the average execution time with different numbers of threads. But quickly I saw that sadly the second method isn’t relevant for what we want to do. Indeed after a quick test I realize that asyncmap isn’t using multi-threads and thus does not speed up anything when its ntasks parameter varies. The resolution time is always almost the same as the single threaded version of the code. First I though that my code was wrong and this behavior came from mistake of mince but later I read more closely the Julia documentation and I noticed this little note in the end which says “Currently, all tasks in Julia are executed in a single OS thread co-operatively. Consequently, asyncmap is beneficial only when the mapping function involves any I/O – disk, network, remote worker invocation, etc.”. So indeed co-routines will not help us (today), but does threads ?

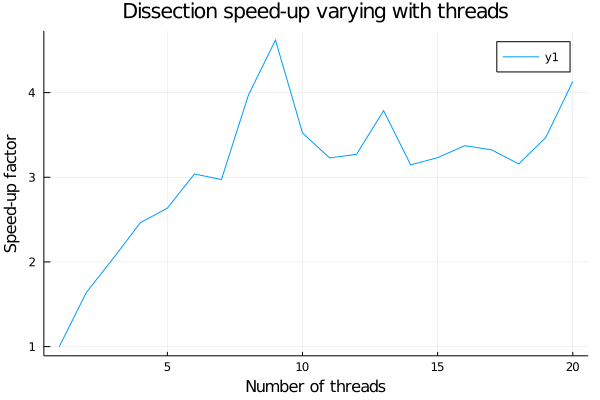
From this little experiment we can deduce that for the usual 20 calls to flow-cutter it’s better to use 9 threads to minimize the run time. Now we can write an MPI version of a wrapper which will call in different processes the dissection function with different seeds to ensure different RNG and therefore different results to compare in order to find the best order. In this wrapper, we need to be careful at different things:
- The MPI script should accept several options to take properly care of hyper parameters like nsample, seed, max_imbalance, etc.
- Change parameters between iteration to improve the tree-width for example: augmenting nsample, round the max_imbalance to increase diversity of separators.
- Manage seeds to ensure every pair (processID, threadID) has a different seed to compare different results.
- Only call it with a number of process that matches with hardware i.e nprocesses * (nthreads=9) <= navailablethreads
- Take as input a duration in seconds to stop in time and give best order/tree-width.
- Add a stop condition in the dissection to return when we get a tw > current_best_tw and thus avoid wasting time for bad orders.
Thanks to the Julia community using MPI is quite accessible with the MPI.jl package which has a clear documentation full of examples. After one afternoon testing the MPI script on the supercomputer Kay from ICHEC, I finally managed to make it run without errors. The results however are not as good as we expected. On the sycamore 53 20 graph, according to the the implementation provided by the authors of the paper, we should get a tree-width around 57 for 30s of computation. Within the same amount of time, (8 processes, 9 threads on 2 nodes of Kay) we poorly reach 94, which is quite far from the expected value. Waiting longer doesn’t seem to help a lot, for 2min so 4 times longer we get 92 so a minor 5%(= (94 – 92) / (94 – 57)) improvement. This bad behavior seems strange. We suspect this to happen because we choose to keep an undirected graph in the core flow-cutter part and not to use the expanded graph.
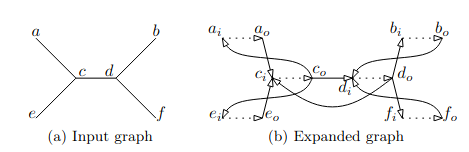
The expanded graph is a technique to map an undirected graph to a directed one. The process involves doubling every node and edge, so we want avoid it. If we use an expanded graph, the forward grow and backward grow will be different and I suppose that’s why that it leads to our weird behavior. My mentor John Brennan also thinks that another issue could be the way we compute the tree-width during the main loop in the dissection part. Indeed, during the implementation, we encountered several difficulties with it. Sometimes it returns a value with a distance of 1 to the value expected by a posteriori computing the tree-width from the order. This difference is mainly due to a formula used to compute size of close cells. This is detailed in this paper from the same authors, which seems true but actually is false in some specific counter examples. We emailed them about these issues, and they answered being aware of this, and said the formula turns wrong only when a sub-optimal separator is chosen. This matches with our practical test cases where with some seed, the RNG picked a wrong separator and we can observe this behavior. The only remaining way to fix this that I saw, is to change the implementation of the separator function in order to create an expanded graph before calling flow-cutter and change the growing method to differentiate forward and backward.
Sadly this Summer of HPC will end this weekend so the timing is too short to try this fix to improve the algorithm. It’s really annoying to end a project like this with an algorithm not fully complete. However I think I did a nice job writing maintainable and readable code, therefore I hope the project will continue or be forked. In the end, this summer was quite exciting with all the things I learned about graph algorithms, MPI, Threads, Julia language and English language as well 🙂

Leave a Reply