A Week Full of Surprises
Man at Work
We arrived at the first day, after a long and peaceful weekend. Lukas went to the beach, but concluded it was too small for him. There are more to look after. Days in Copenhagen can be tricky, the morning starts with a cold refresh, and then becomes hotter (by local means).
But this is not the point, we came full of tasks and ready for actual work.
The first goal was to profile our algorithms on the cluster with different compilers and input data. To accomplish this task we first added support to run every algorithm independently so results are not biased.
The local cluster is composed of several hundred of CPU cores (AMD Opteron 6272) and utilizes Slurm for batch backend. Each node has supporting GPU device (AMD Radeon HD 7850) for accelerated performance. Quite cool.
For those who like astonishing raw numbers, this GPU is made of 2.8 Billion transistors, and capable of 1.76 TFLOPS (Trillion Floating Point Operations per Second).
The cluster architecture is very similar like the Titan, but Titan has much more nodes. This cluster has only 9 nodes but their names are very interesting. They are Sashi, Pria, Anoop, Gheet, Poonam, Uma, Sandeep, Nabendu and Manjula. Do you know these names? Maybe another name can help you, all data storages have name Apu_number. Yes, now here we are all Apu’s family from The Simpsons. Evereythink what belongs to the cluster has name from that famous cartoons.
Back to reality …
The compilers set includes, g++ 4.6.3 (Cluster), g++ 4.7.2 (Us) and also Intel 13.1.1. Our project were build against each and ran with input of 10000 particles, clustered into 10 centers.
To average performance results, the processes were ran on 4 different nodes, with same input.
Now several surprises were expecting us:
1. The Lloyd algorithm, performed best, in all scenarios (it was expected to be worst).
2. It seem that for some algorithms AMD processors are slower twice than Intel. Thanks to Lukas who was testing.
Further investigation led us to understand that Lloyd, wasn’t implemented in naive way, but used an advanced KD-Tree data structure. This provides a significant performance boost.
The newer g++ compiler gave slightly better results (2.5-5%), but Intel was better with 10% faster than all.
Still, this is not the domain we are looking for, improvement by some percent leaves Lukas agnostic. He is looking for the magnitude factors. But several goals are already met.
Original Python implementation
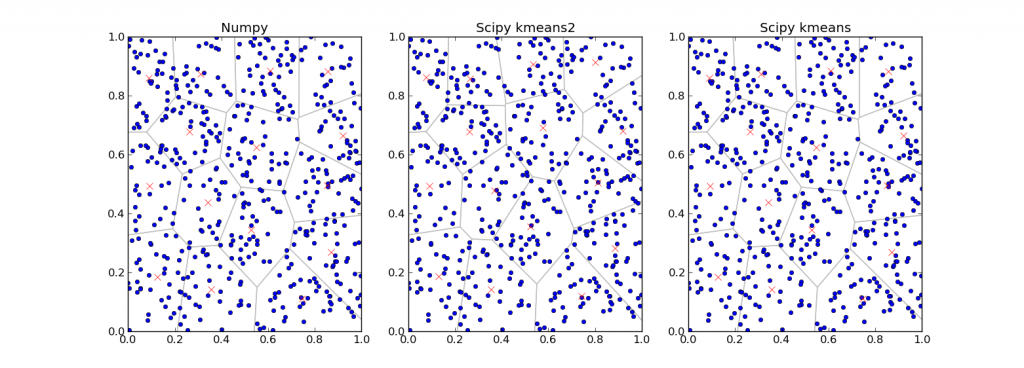
As we mentioned in last post we got a paper, a Bachelor Thesis, prepared by a local student and summarizing previous work on topic, including implementations written in Python. So we wanted to compare results for C++ code with the python code. In the python code there was prepared some benchmark for different implementations using OpenCL, CUDA, Numpy and Scipy. It seemed to be easy, just run the benchmark and compare runtimes for each implementation. But it wasn’t, because the output of the benchmark contained a lot of useless informations (listing of variables, listing of memory usage for application) and just a few useful informations like listing of current implementation but without runtime. So to understand and to correct the python scripts took half a day. We had to also set the same initial and stopping conditions like in our C implementation, what meant to rewrite some functions in Python code and add some new. Fortunately, when you want rewrite some code from C to Python, all work consist in copying the code and erasing useless syntax.
Finally, we had desired data for comparing with C code. When we will consider only Numpy implementation, the original one written by author of the paper, it was defeated by our C implementation. That is because of the C implementation using very effective tree structure called KdTree (https://en.wikipedia.org/wiki/K-d_tree). Python build-in implementation Scipy k-means was on similar level.
Vectorization Support (SIMD with SSE/AVX)
K-Means algorithms of different flavors do basically the same, compare distances between a point and its allocated cluster/center. The core functionality being used is thus computing distances, in Euclidean spaces (called L1 or L2 norms).
After we gained some insight into the libraries, we thought it could be a good opportunity to introduce vector operations to compute distances, clearly an operator that repeats itself most of the time during computation. Most implementation do not support that as they are meant to run on various CPU architectures and supported instructions vary.
The idea to use first vector operations, is to build a hierarchy of optimizations, from the more trivial to advanced (OpenMP multi-core and MPI distributed).
To use SIMD operations, one can program in assembly code (No), use specific libraries or intrinsic functions provided by the compiler of choice. the drawback of using libraries, is that they are generic, and usually introduce new datatypes as well. So we decided to move on with intrinsics, support by Intel and GNU compilers (gcc/g++).
It took no more than few hours (including bugs) to have support for core routines in the algorithm with SSE/AVX instructions. We decided to target our new platforms, and not support legacy systems, therefore, utilizing the latest AVX instructions we can on our cluster and development system.
For some reason, the functionality exposed through SIMD operations on Intel processors is not complete for floating point operations – there are many useful operations that needs to be built by several instructions, where there exists an integer counterpart with a single command. Not to mention that single precision gets more support than double. But we are not the designers (Integers are good for general text/image processing).
Our final solution was implemented by operating on 2 items of double precision at each stage. Just for simplicity, it is easier to handle odd length arrays without sacrificing too much performance, and as an educational tool. Once we’ll implement the final version, the code will be specific and highly tuned (counting each clock cycle and latency!)
As a side note, to demonstrate why vector operations are powerful, we can consider the stopping condition for the algorithm: if center are changing only by little (~0.1%) between iterations, then we are due.
The original algorithm was using L1-norm, clearly for performance reasons (not squaring any numbers, just normal arithmetic). But with SIMD, the way to implement the L1-norm had to use dot product (“lack” of other options), but good since this is a very fast instruction. From the CPU perspective, computing dot product between v1 or v2 is the same as v1 with v1. So, we could also test the algorithm when the stopping condition is computing L2-norm.
Two “surprises”: First, we reduced some overhead instructions required for L1-norm (less instructions, more throughput). Second, L2 is more accurate than L1, so the algorithm stops faster (less iterations) and is more accurate. All, in the same price (or even cheaper). In higher dimensions (such as 6), this yields performance improvement of over 50% measured.
Final Notes
Our algorithm of choice performs in C++ better than all other versions, even those implemented in python using NumPy/SciPy (tuned & parallel) and also compared with OpenCL performance on the GPU.
We cannot be more satisfied than this, with our plans forward toward next week.
Future Work
During the next week we want to do more detail profiling on cluster and also continue with vectorization and another optimization of C++ code. Finally, we should get a realistic dataset from Photon-plasma project. Next goal will be to try visualize 6-dimensional data to recognize which results are better.


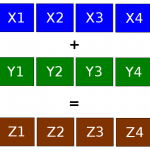

[…] Moti and I contended with vectorization for a few days. As mentioned in one of our previous posts, we decided to implement SIMD vectorization using Intel SSE/AVX instructions into our C/C++ […]