The end of the beginning!

We have come to the end of my project! I cannot believe how quickly this internship has gone and I am so grateful for the opportunity. I hope you enjoy this final blog post of mine as I explain the last tasks undertaken in my final – and potentially the busiest – two weeks.
With my project now finished, I am pleased to be able to provide details of the final steps taken. To summarise, I worked with four variants of the K-Ras4b protein: K-Ras4b wild-types each bound to GDP and GTP molecules, and K-Ras4b G12D mutants each bound to GDP and GTP molecules. As a recap from a previous post, wild-type proteins refer to the unmutated, normal protein. Contrastingly, the G12D refers to a specific mutation that has occured in the protein: the glycine amino acid in position 12 along the amino acid chain has mutated, becoming aspartic acid. GTP and GDP are known to bind to K-Ras4b. When bound to GTP, K-Ras4b is active, meaning that cell proliferation can occur. This refers to an increase in the number of cells as a result of cell growth and division. When K-Ras4b is bound to GDP, however, it is inactive so cell proliferation terminates. The G12D mutation in K-Ras4b means that when bound to GTP, it cannot undergo hydrolysis. This is the process in which GTP becomems GDP and effectively acts as a switch to turn off the cell growth and division. As a result, since the growth and division of the cells cannot stop, this can lead to tumour growth! My project involved studying this mutation in particular and running molecular dynamics simulations in order to investigate the changes within the protein throughout its trajectory. The resulting trajectory for the mutated G12D K-Ras4b protein, bound to the activating GTP, is shown in the animation below. This was visualised using VMD (a visual molecular dynamics program). The part of the structure that is shown using a different representation (refer to previous posts to explain the various drawing methods in VMD) is the aspartic acid molecule in position 12 along the amino acid chain. This is present since the G12D mutated K-Ras4b is displayed, so the glycine in position 12 has changed to aspartic acid, as explained previously.
For the molecular dynamic simulation processes that were carried out, ‘jobs’ were to be submitted to the ARIS supercomputer. These are files that essentially tell the supercomputer how to process the information it has been given. Unfortunately, during my project, I sent some incorrect jobs to the supercomputer. This did not affect the project as a whole but just delayed the work schedule that was in place by a couple of days. One of the very important lessons that this internship has taught me is that projects like this are a long process that will sometimes include slip ups but if you keep working hard, you will get there. It has taught me that I can often learn lessons very deeply after having made a mistake since this requires me to investigate what went wrong. This then teaches me more about the systems used than if it had all gone correctly the first time! I suppose that this is what an internship is all about: learning new information and methods, making a few mistakes, learning from them and then developing your knowledge base as you progress. This internship has also, importantly, shown me how a working lab runs and how to work both individually and also as a member of the lab, offering and asking for guidance from peers.
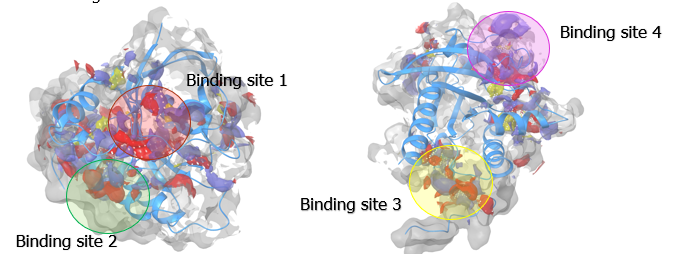
Drugs are small molecules that can block proteins – which have cavities where drugs can bind. Within a protein’s structure, they may have a burried binding site, which is where a molecule can bind to the protein. A small drug molecule can be found or specifically designed in order to fit this binding site. Therefore, a crucial step in the project’s process was identifying binding sites on the four K-Ras4b proteins studied. After running the molecular dynamics simulations, this binding site identification was performed and binding sites were found. These are shown in the image below for the mutated K-Ras4b protein bound to the GTP molecule. This process was repeated for all four protein variants.
The very last tasks of my internship included creating a video that explained and summarised my project! This can be seen on YouTube by following the link here . I would love to hear what you thought of it and receive your feedback! Moreover, on the last day of my intership, the 28th of August, I gave a 20 minute presentation to my colleagues here at the Biomedical Research Foundation of Athens. We can all be seen in the image below.

My final thoughts on the internship are that I have learnt so much and am so grateful for being accepted onto this programme as I have gained so much experience. I will take the skills learnt with me into the future as I know that they will be helpful in the upcoming projects I undertake – hence the naming of the title: the end of the beginning. Even though my internship is now over, this is still the start of my journey and I look forward to my future endeavours. Moreover, I have met so many wonderful people from this internship and I wish them all the best success in the future. Thank you so much for reading my blog posts and I can now say thank you for reading my last blog post!

Dear Rebecca, as an organic chemist at Oxford University I have really enjoyed learning about your progress and these final stages of the project. You have taught me about the great opportunities offered by PRACE – I wish I had done something similar prior to starting my PhD. I hope you continue to research for effective alternatives to animal testing. Good luck!
Hello Harry. Thank you so much for your comment! I really appreciate your support! I’m so pleased to hear that you have enjoyed learning about the project I completed! Yes, I am so pleased to have learnt about the PRACE opportunities! Thank you very much!