DNA and Quantum Computers
DNA sequence alignment algorithms use heuristics methods in order to cope with the high volume of data that is passed to them. These approaches, usually run on super computing clusters, take days to finish due to the amount of data, and usually with an approximate solution. This permissiveness with the solution makes these algorithms possible to work with quantum processors. (See more in detail information here.)
In our project, we mainly focused on the string encoding and comparison. For the alignment, we decided to work with a naïve approach, but of course, further improvements could be done using a better algorithm. Regarding the original project idea, I want to remark that we ended working with Qiskit from IBM instead of Forest from Rigetti, so that would be a small change regarding the quantum computing software platform.
Of course, before start with the encoding and comparison, it was necessary to understand some quantum concepts and how quantum computers work. If you are interested in that I really recommend reading Benedict’s post, where he explains some basic concepts that we have to understand before getting hands-on.
Encoding
Getting back to what we have been doing this past week the first challenge we face was trying to encode the strings using Quantum Registers. For this task, we divided the information between the position of the element in the string and information about the element itself.
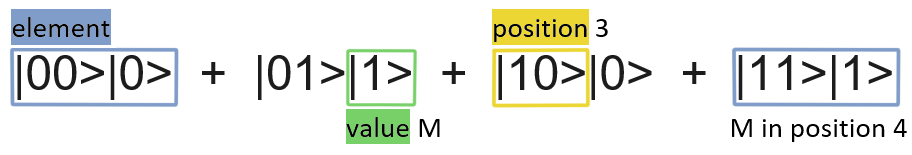
Both pieces of information need to be translated into binary. For positions 0 for the first one, 1 for the second, 10 for the third, … and so on. For the values of the elements, we need to know how many different values are in the string. If there are just two values, one would be represented by 0 and the other by one, if there are four: 0, 1, 10, 11 would be the binary versions of each of the different values.
So, let’s put an example: we want to encode the string ‘-M-M’. We have 4 positions to represent 2 different values. If we use 0 to encode for – and 1 for M:
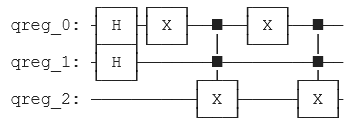
We applied the corresponding operations to encode the strings in a Quantum Circuit. To save memory, we used some properties of quantum computing, such as superposition.
Comparison
Regarding the string comparison, we base that on a simple idea. We initialized all circuits at 0 for every Quantum Register. Once we have applied the operations to encode the string, if we applied the same operations but in reverse order, the initial state of the circuit will be obtained, with all registers at 0.
Therefore, when we want to compare 2 strings, we are going to add the operations of the second string in reverse. And then, we will check for the Quantum Registers, if all are at 0, that would mean that the 2 strings are identical. The more different the strings are, the more 1 will be found in the quantum circuit. For example, if we compare ‘—-’ to ‘MMMM’ we will obtain that all registers are at 1.
What is left to do?
More or less we resolved these two challenges for DNA sequences, and in a way that can be more or less general for strings with different information. Now, we are left to do the final implementation and measure performance.