A Final Chapter

Hello everybody!
Welcome to my third blog post, where I will be talking about my expirience with Summer of High Performance Computing (SoHPC). Once again my name is Ioannis Savvidis and if you would like to get to know me a little better and see what we have done up until now go check my first post which is an introductory blog of who Ioannis is, and my second post where I write a bit more about the project i work.
So in my last post I explained how we managed to find a working implementation of sparse BLAS library, and we successfully implemented it to our existing code. After some testing to a methotrexate molecule, the results that came back were not looking good. The new code with the sparse BLAS implementation was slower than the old code.
In order for our code to shine we knew that we have to go for bigger molecules where the matrices are even more sparse. That’s how started testing to 5 different alkanes molecules by running the old and new code and checking if the run time is getting better or even if we are having faster run times. The alkenes that we used as i said in my second post were C6H14, C12H26, C18H38, C24H50, C30H62, C36H74. A schematic together with methotrexate can be seen on the left.
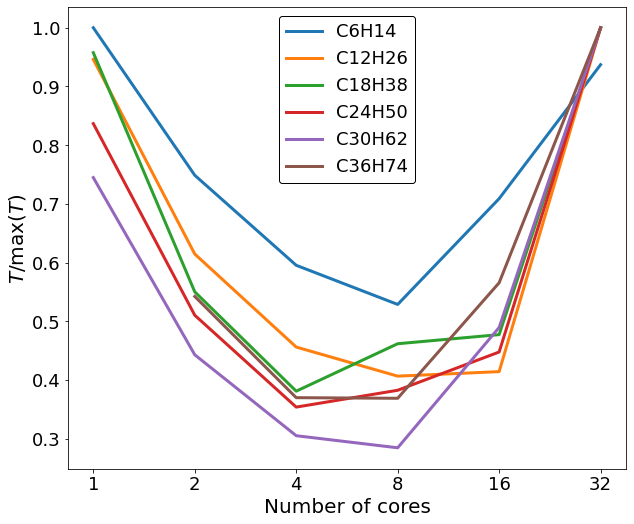
At first a single node of a supercomputer was used exclusively to do our testings. Each node of the supercomputer has 4 CPUS @ 8 cores. So we tested the timings of the codes for different number of cores, specifically 1, 2, 4, 8, 16, 32. From our testing alkane molecules, almost all of them had faster run times at either 4 or 8 core with the majority of them at 8 cores, while the slowest timing were coming in the 32 cores runs.
From all the data we collected, we were able to determine that for C36 why had an average of 4.2% improvement of runtime on our new code.
After that point and with more nodes to our disposal, we tried an implementation of parallelism by testing how the run times scale with 1 to 4 nodes that are set at 4 and 8 cores. Unfortunately, the timings weren’t consistent and further troubleshooting is needed to be done. Finally, on our last week on the project we increased the number of basis set function for C30 and C36 molecules and tested the new code. Again at the end we run to some numerical errors and we don’t have the time to run more tests.
That concluded our journey with SoHPC. I want to say that I’m really happy with the outcome of our project and i feel really greatfull for the opportunity of participating on this year’s programme. I learned a lot of new things under the guidance of our mentor Dr. Jan Simunek and help from my colleague Eduard Zurka. Also, big shout out to Dr. Leon Kos and PRACE for organizing this programme and everyone that helped us from the training week until the end. Ultimately, I want to say to everyone that reads this post and has an interest in HPC or wants to learn more about HPC and has an interest in computational materials science/chemistry, to apply to next year’s SoHPC.
Thanks for reading my blogs! Be safe!

Leave a Reply