Can a theoretical physicist love biochemistry? (Spoiler: YES!)
After completing the training, my final project has now begun!
DAY 37/60
After the initial week of training in Italy and the month of training in Greece, my final project has begun! I am so grateful for this in-depth training period as it taught me an array of knowledge ranging from molecular dynamics simulations to the in depth biology and chemistry that explains some of the science governing them. As a theoretical physicist, this has been very new territory for me. However, it has shown me that my skills acquired from physics have been transferrable. In addition, for the past couple of years I have been intrigued by biophysics and this internship has been inspiring and has assured me that introducing biophysics modules into my master’s degree next year would be of great benefit to me. If they are anything like this internship, then I know I will find them extremely engaging.

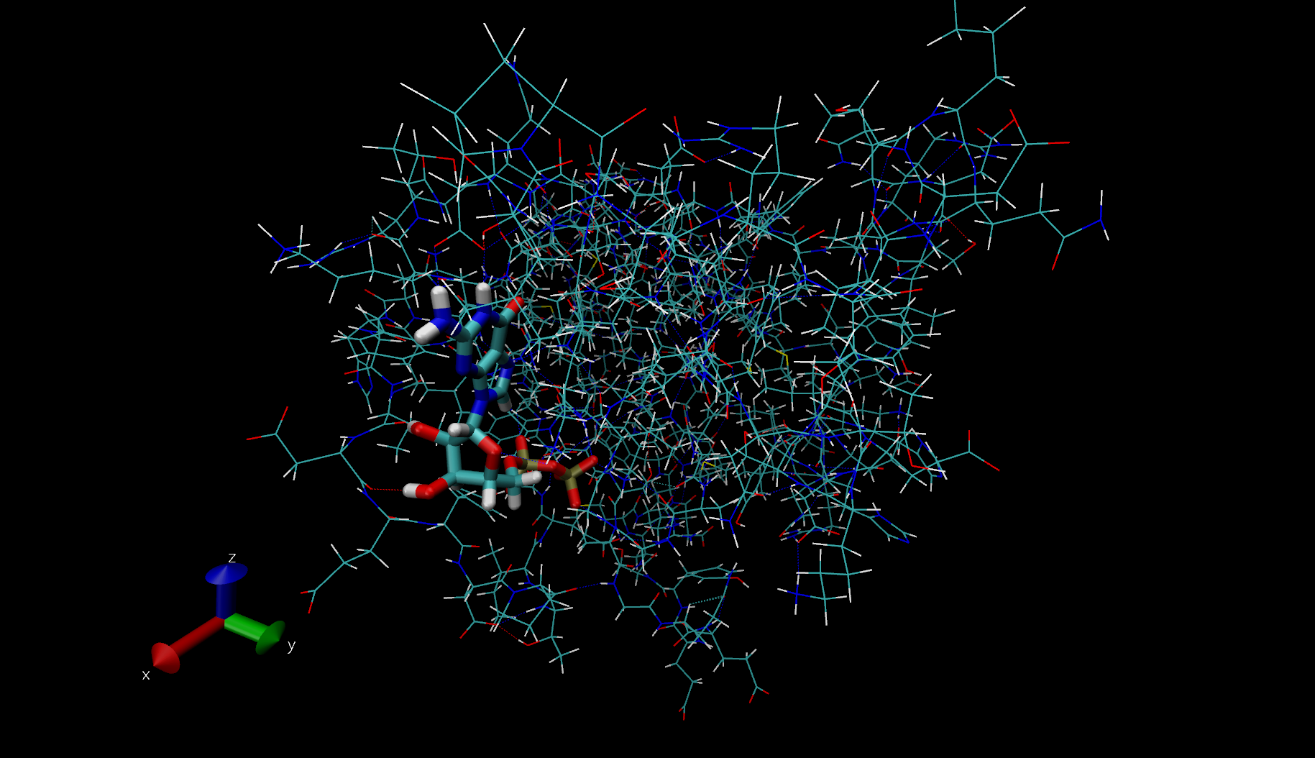
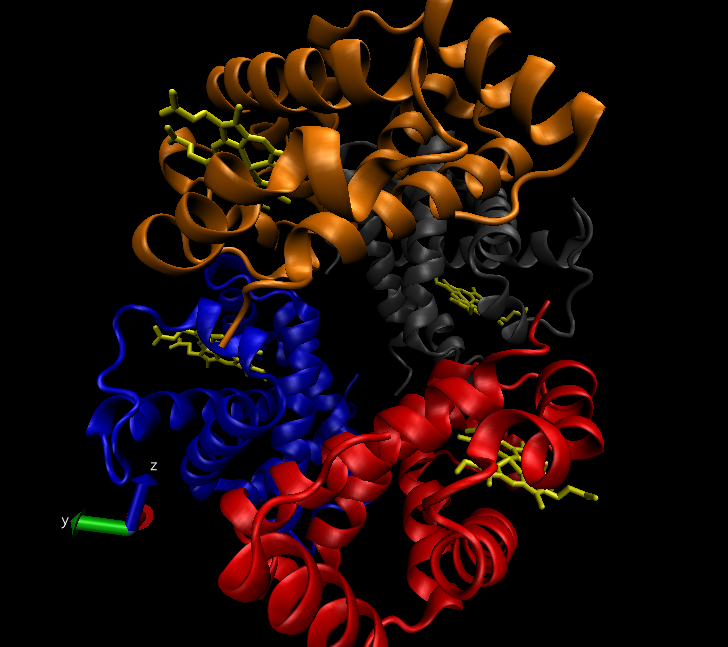
As a recap from earlier on in the internship, a Nanoscale Molecular Dynamics (or as some call it, Not Another Molecular Dynamics program), (NAMD) tutorial was completed, after writing a report on the inter- and intramolecular bonds in proteins and protein-ligand complexes. Last week, the work I had done for these tasks were reviewed and after meetings with my mentor, helpful updates were included, allowing my work and my understanding of the science to develop further. I also completed another two tutorials that aided my understanding of the process behind drug design and the biochemistry that governs the interactions and bonds in proteins. For example, this included studying haemoglobin, a protein in red blood cells. The protein data bank (pdb) file, which is needed in order to visualise the structure in VMD ,a visual molecular dynamics program, was downloaded from the RCSB (Research Collaboration for Structural Bioinformatics) website. This protein is shown below. A range of other proteins I investigated are also shown in the following images. Some of the images represent the same protein but are displayed using different drawing modes. These different ways to view the same structure have shown to be very useful throughout my project as depending on the reasoning for looking at the protein, the most suitable drawing type to use can be determined. For example, if the general structure of the protein is to be examined, the ‘NewCartoon’ drawing method is helpful. On the other hand, to see the individual bonds, the ‘licorice’ drawing method may be the most appropriate.
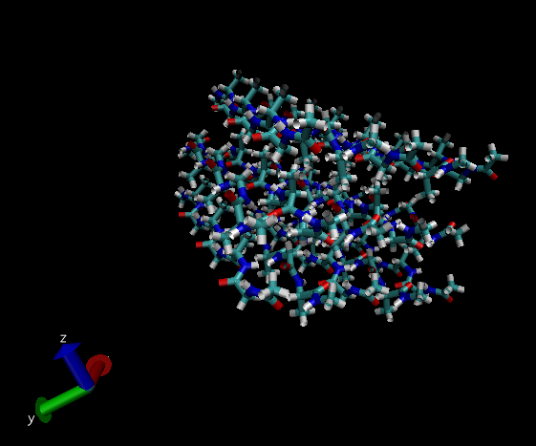
This image shows beta sheets of model silk fibroin peptides using the ‘licorice’ drawing method. 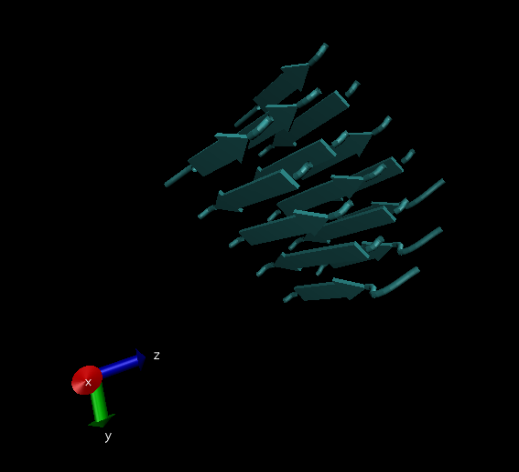
This image shows the same protein as the previous image but using the ‘NewCartoon’ drawing method.
My favourite parts of this intership so far have been visualising the proteins and the final simulations at the end of each step in the process. I find the capabilities of these molecular dynamic simulations and supercomputers so incredibly fascinating and I feel so lucky to be able to learn about how to utilise them. I am so excited to see what the future brings, as I hope to one day join the scientists working to use these simulations, and others like it, to reduce animal testing. I am so pleased to be working on a project that is teaching me essential knowledge required for this field of study.
After finishing these tutorials, I completed a Glide and a SiteMap tutorial- the latter of which I will expand on now. The location that a potential drug could bind to a protein is usually known if its structure is known. However, in some cases, the binding site of a protein is unknown, making it hard to know the types of drugs that could bind to it. Therefore, computational studies can be used to help find these potential sites without much prior knowledge of the protein structure. Initially, a couple of regions on the protein’s surface, called ‘sites’, are identified as potentially promising. These are located using a grid of points called ‘site points’. Various further stages entail which all help ensure the best chance of successfully finding a good binding site. This information is provided to Maestro in order to visualise the process. (Source: SiteMap User Manual, Schrodinger Press, 2009)
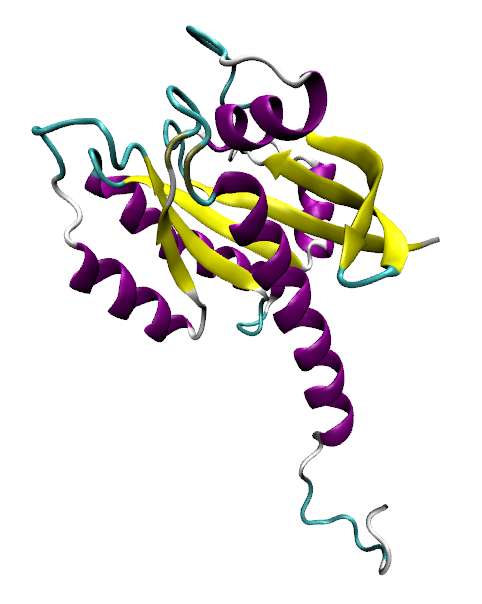
The very first task of my final project was to write a document explaining the functional domains, the mechanism of action and how the mutation affects both of these for the K-Ras4b protein. I found this task so beneficial to clarify both my understandings and mis-understandings of these processes and ensured that I was clear about the properties of this protein. The NAMD tutorial, completed previously, was almost like a trial run for me. The tutorial focussed on the protein ‘lysozyme’ and took me through multiple steps such as minimisation, heating and equilibration. After learning about these processes, along with the other tutorials, I am now ready to start my final and main project. I will be performing similar minimisation, heating and equilibration steps (just to name a few) on K-RAS4b proteins (one of which is shown below) which include both the wild type (referring to the unmutated state) and the G12D mutated protein, meaning that the amino acid in position 12 has mutated from a glycine to become an aspartic acid. I will then use SiteMap and Normal mode analysis to do binding site identification in order to determine whether any cavities exist on the proteins in question.
Still to come… As the final few weeks approach, there may just be a few more tasks to complete (as well as the project itself) however they may be the most crucial of all! There will be one more blog post by me in two weeks time which will summarise my project, give some more details about how it was carried out and finally, a summary of the results obtained- I promise to make sure it includes plenty of really interesting videos of the simulations and photos to accompany them! I am also in the process of creating a 5-minute video to explain my project from beginning to end and also a final report that will be submitted to PRACE. Moreover, I will be delivering a 20 minute presentation and then showing my video to my colleagues at BRFAA which will finish off my internship completely. I hope you can follow me on the remainder of my journey! I would also love to hear if you have done any similar projects this summer or if you have an interest in something in the same area of study! I’d love to hear about all of your thoughts in the comments below!

This sounds so interesting! It’s great to see that different areas of the sciences can be applied to this field which don’t necessarily relate, and how you’ve applied your own knowledge. I’m excited to see how your project ends and what you’ve learnt!
Thank you so much Emily! Yes, definitely! It has been so interesting to see how biology, chemistry, physics and computing have all come together to create this vital field of drug design! Thank you – I look forward to keeping you updated with my progress.
This sounds amazing! I’ve been looking forward (since my last comment) to hearing more about your final project. I’ve always wanted to use a supercomputer! This would be an amazing tool for my area of research – drug synthesis! What a brilliant opportunity from PRACE!
Thank you so much for keeping a look out for my blog posts!
Me too – I have always wanted to use a supercomputer and feel very fortunate that the PRACE Summer of HPC has allowed me to do this!
That sounds excellent! I hope it goes well.
It looks like you are progressing very well on your project- especially considering this is a new area of study for you since you are more used to Physics! It looks like you have learnt so much!
Thank you so much! Yes, biochemistry is a new area for me but I am really enjoying using the skills acquired from my physics degree to transfer them to this new field! Yes, I really have learnt so much new information, ranging from how to use molecular dynamics simulations to the structures of K-Ras, the protein studied for my project.
Loving the visual representations of the work you are doing, so often a lot of physics is just lines of code which can be boring to read and even more difficult to understand! Sounds like the project is going really well, are you going to post the 5 minute video explaining your project in full on here because I would love to watch it?
Thank you so much! Yes, I love seeing the visual representations too – they fascinate me so much!
Thank you, I have learnt so much so far and am still learning new things every day.
I would love to post the 5-minute video on here! It will definitely be on YouTube so if I am not able to post it on here, I will be sure to post the link to YouTube.
Thank you for your comment!