Indeed, as the summer comes to an end I realize I had a lot of rain – both here in Scotland and on my computer screen. My project was to develop a weather visualisation application which will be used at outreach events, so lets take a look how that went.
During the project I have developed a application which will be used for outreach, to explain simulations, HPC and parallelism. At the beginning of the project the idea was to do just a weather visualisation demo, but after a few discussions with my mentor we realised that this has a lot of potential. So apart from the clouds and rain, I decided to expand the project and display some other interesting information about the performance and how parallelism is done. Also, the application was meant to be used just at outreach events, but with all its parts, it is perfectly suited to use as an education tool, which some people already plan on doing. It can be used in regular classes or in training courses both for young meteorologist and other scientist that want to use HPC and understand how it works.

A screenshot of the application
Users can select input parameters for the simulation. Of course, meteorologist use much more input parameters than the 5 we have here: wind power, atmospheric pressure, temperature, time of year and water level. They are chosen for the purposes of outreach, parameters everyone knows and can understand. Time of year translates into the force that pulls the water from the sea into the atmosphere. With water level we determine the amount of water that is available at the bottom of the atmosphere. There are some other options which do not influence the outcome of the simulation, but the performance of it. This may be of interest to computer scientist or beginners in the HPC field. Here we can select the number of cores per node the simulation will run on, but also the way the decomposition will be done. The simulation uses a 2-D decomposition technique to split up the workload amongst the nodes, although the simulated space is in 3-D. Each core will then get some piece of the atmosphere to work on, which will have a custom width and depth, but will take all of the height, ranging from the ground to the highest level of atmosphere. We set the number of processors that will split up the workload in both width and depth. We can also set one of two solver techniques: Incremental or Fast Fourier Transformation(FFT) and see how it affects performance.
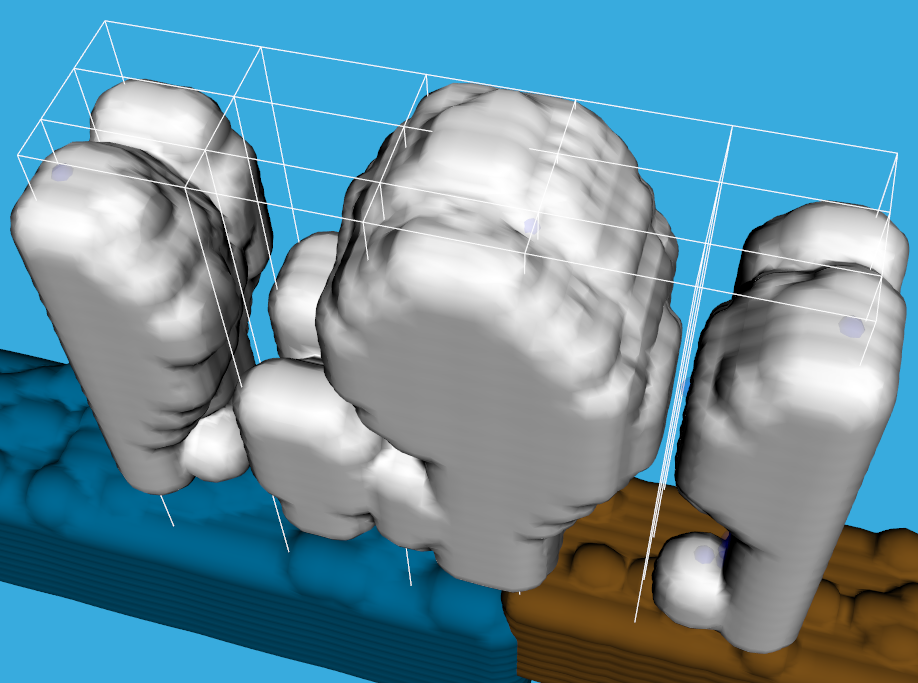
A better view of the decomposition grid
The cores in HPC systems usually have to communicate and share results and data, this communication often takes a big part out of the overall time. We agreed that it would be nice to see how different decompositions affect the communication. A visualisation is generated each time Wee Archie generates a data file. Clouds and rain are rendered, and we can see how the cloud forms and moves. If we got the input parameters right, rain will accumulate around the clouds and start falling. A gamification has also been included, if enough rain falls crops will rise out of the land, but too much rain can destroy the crops. A grid showing the decomposition is also rendered so we can see how the atmosphere is split up amongst the processes. The other part of the visualisation is the plot at the bottom part. For each core running the simulation, there are two bars; a green one showing computation time out of overall time, and a red one showing communication time out of overall time. We can then easily compare how much of the time is spent in communication and how much doing computation. Of course, it is of interest to tweak the settings so that we have low communication time and high computation time. Another performance measure appears in the upper left corner, which shows how much simulation seconds are computed in one real time second.

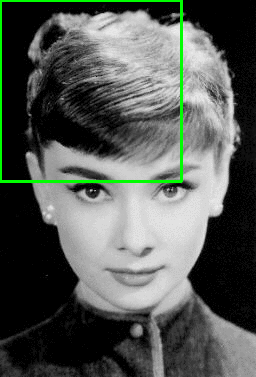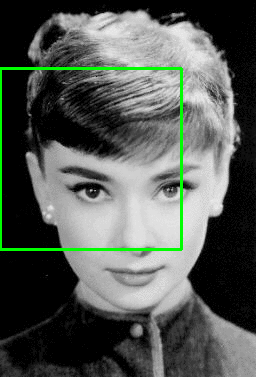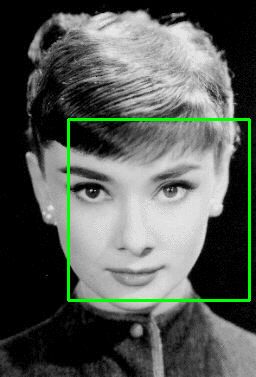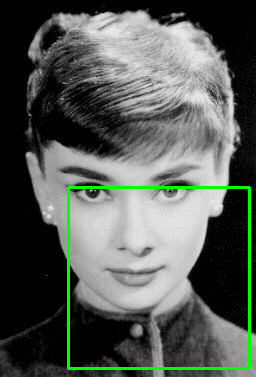
Light showers above the coast
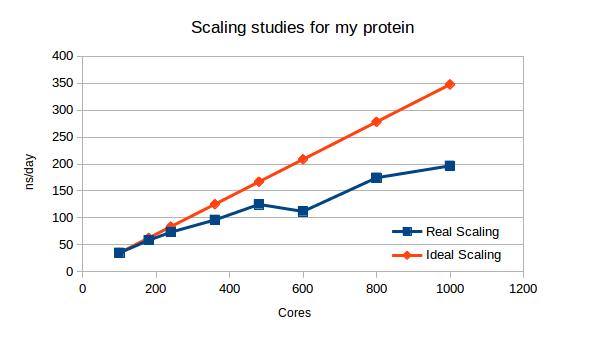
Most scientist that are starting to use HPC are not aware of the trade-offs they have to make. Some methods in the simulations may be give the same results, but the performance is different. This application can give them an idea that performance in HPC depends on a lot of things. The decomposition grid is a perfect way to show how the data and the workload is distributed amongst the cores. With different settings and performance results we can easily show that just running simulations on a large number of cores is not enough and that we have to find the best way to distribute the workload.













 Anyway, the next step was to make the code parallel. I used the OpenMP library for this. The library provides a simple framework for letting the code run on multiple cores on one computational node. With my supervisor, we decided to use OpenMP for its simplicity, and also for the fact that the parallelisation (uf …) of the code is only possible over the space domain, and not the time domain. In other words, all threads must
Anyway, the next step was to make the code parallel. I used the OpenMP library for this. The library provides a simple framework for letting the code run on multiple cores on one computational node. With my supervisor, we decided to use OpenMP for its simplicity, and also for the fact that the parallelisation (uf …) of the code is only possible over the space domain, and not the time domain. In other words, all threads must 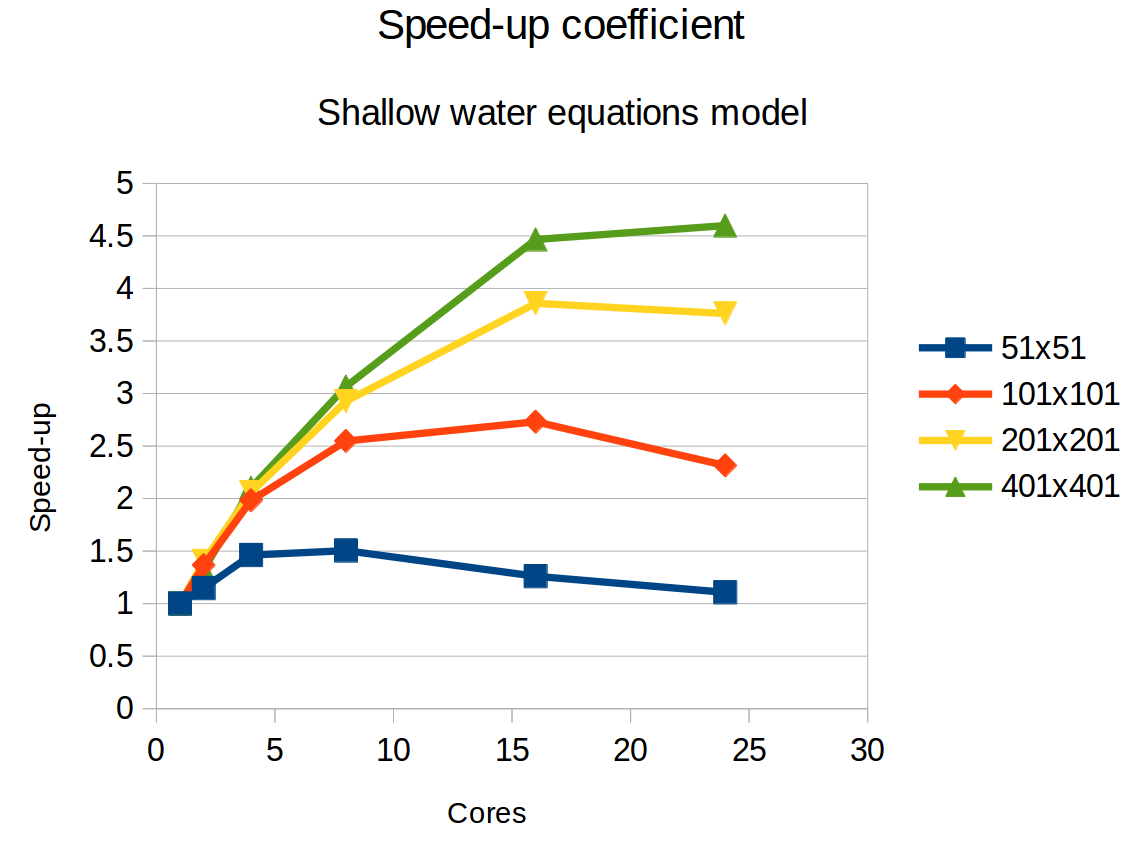




































































 The first day was in the name of familiarizing with
The first day was in the name of familiarizing with