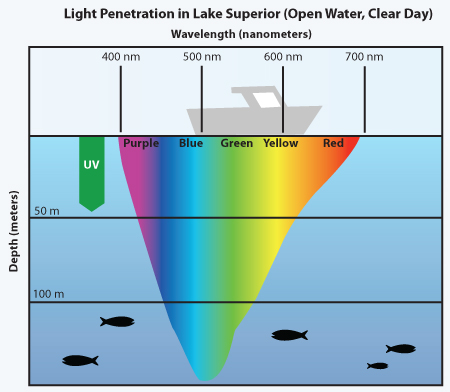
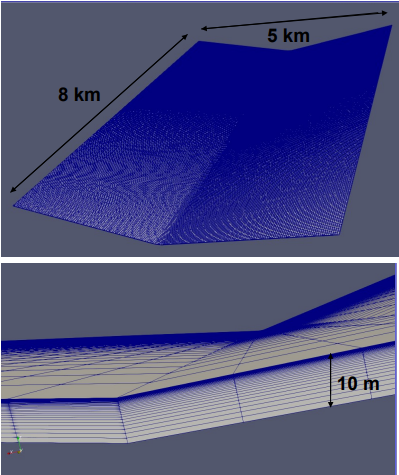
Computational Fluid Dynamics (CFD) is a branch of fluid mechanics that approximates the time-dependent partial differential equations numerically (PDE) that dictate fluid flows, the Navier-Stokes equations. The Navier-Stokes equations describe phenomena we encounter in our daily lives. Ranging from the description of today’s weather to the flow of blood in a dialysis machine and from the flow inside natural gas pipes to the complex flow around the wing of an airplane. Despite their broad applicability, the proof that there is always a solution to them remains one of the Clay Institute Millennium Prize Problems.

The numerical approximation of the Navier-Stokes equations is performed using a wide variety of methods. Most of them require a computational mesh, for example, the Finite Volume Method, the Finite Element Method, or the Finite Difference Method. On the other hand, some other methods do not require a mesh, such as the Smoothed Particle Hydrodynamics or Lattice Boltzmann Methods. Both types of methods greatly benefit from parallel computing, utilizing strong (speedup with fixed problem size) or weak (speedup with increasing problem size) scaling.
Finite Element Method
In this project, I utilized the Finite Element Method (FEM). This method is complex from a mathematical standpoint, so I will try to summarize the central concept without getting into further details. The main idea is to split the computational domain into tiny geometrical shapes (mostly triangles and rectangles for 2D or tetrahedra and hexahedra for 3D) and using polynomial functions, called base functions, we approximate the flow field inside each element. The final solution is considered to be a linear combination of these functions. Thus the problem becomes the determination of the base functions coefficients (or degrees of freedom), leading to a simple linear system of equations, which is solved using direct or iterative solvers. The final system is obtained using the Galerkin method, multiplying by weighting functions (usually the same as base functions) and integrating the original equations (strong form), leading to the so-called weak formulation.
So, where can HPC contribute? We can use HPC systems to use even smaller temporal and spatial discretization, leading to huge matrix equations that efficient parallel iterative or direct solvers can solve.
FEniCSx
The next question is how can create these weak formulations, choose the base functions, construct the mesh, etc. The answer is using FEniCSx! FEniCSx is the newest version of the FEniCS code (now considered legacy code), a popular open-source computing platform for solving partial differential equations (PDEs). FEniCSx enables users to quickly translate scientific models into efficient finite element code using automatic code generation. It also provides high-level Python and C++ interfaces, allowing users to rapidly prototype their applications and then run their simulations on various platforms ranging from laptops to high-performance clusters using MPI. The main FEniCSx components are:
- DOLFINx is the high-performance C++ backend of FEniCSx, which contains structures such as meshes, function spaces, and functions. DOLFINx also has compute-intensive functions such as finite element assembly and mesh refinement algorithms. It also provides an interface to linear algebra solvers and data structures, such as PETSc.
- UFL (Unified Form Language) is the language in which we can define variational forms. It provides an interface for choosing finite element spaces and defining expressions for variational forms in a notation close to mathematical notation. Then, these can then be interpreted by FFCx to generate low-level code.
- FFCx is a compiler for finite element variational forms. From a high-level description of the form in the Unified Form Language (UFL), it generates efficient low-level C code that can be used to assemble the corresponding discrete operator (tensor).
- Basix is the finite element backend of FEniCSx and is responsible for generating finite element basis functions.
So during this project, I developed a solver for the Navier-Stokes equations in FEniCSx. In my next post, I will describe the process of obtaining the weak formulation, the test case I used, and my results. Stay tuned!