As I explained in previous posts, I’m following a transfer learning approach to training the model that I will use to detect action units on faces. This means that I’m going to be taking an existing network, removing a few layers and adding new custom ones on top that will serve as a classifier. This speeds up training considerably as you don’t have to build a feature extractor from scratch. However, not fully understanding how a network behaves can make choosing the hyperparameters for the training a hit-or-miss process. This post intends to walk you through my exploring journey in the hyperparameter realm as I explain how and why different training parameters gave me different results, and how I achieved the sweet spot that gave me the model I deployed to the final solution.

The main problem that I faced throughout the entirety of the training process was overfitting. Usually, when a deep learning model is trained on a dataset, that dataset is first partitioned into two: the training split and the validation split. As their names imply, the training split is used to train the model and the validation split is used to test or validate the performance of that model. The reason why we save a portion of the data for validation is that we want to make sure that the model has learnt a general interpretation of the data, instead of memorizing the samples. Memorization happens because a training session involves a model being fed the training data over and over again. So even if we’re seeing an increase of accuracy in the predictions that our model makes on the training set, we don’t know if this is because the model learnt the underlying pattern that we want it to learn, because the model memorized the individual images or because the model is basing its inference on a bias that we weren’t even aware of. This is why cross-checking how the model is doing using a partition of the dataset that the model hasn’t seen yet is so important
Phew, okay. So I chose a set of hyperparameters that I eyeballed to be a good starting point. These hyperparameters that I’m talking about are the learning rate –how fast the model adjusts its weights as an attempt to improve accuracy, the batch size –the number of images that I’m feeding the model at a time, data augmentations –I will go into more detail below, and the topology of the classifier. The data augmentation and the topology of the classifier are not usually included as hyperparameters, but I am for the sake of simplicity.
To begin with, I started freezing the base layers of the network so only the classifier on top would train (as the base layers were already trained to detect image features). I started with a few dense layers (layers of fully connected neurons, in a way like a matrix multiplication) of a size of around 100 each. However, this wasn’t enough. The network was having big trouble generalizing as the accuracy wasn’t improving much. I thought that despite having a set of hopefully useful features extracted by the base layers, the classifier wasn’t complex enough. I then decided to increase the size of these layers.
Larger dense layers started giving better results –it’s worth noticing that increasing the size of the classifier increases the training and the inference time. However, when I reached a size of around 1000 per layer, the training wouldn’t converge anymore. By this, I mean that I would leave the training running for quite a few of epochs (iterations over the entire dataset), but no apparent improvement in the accuracy was shown.
This got me wondering that there might be another hyperparameter that I had to tune in order to continue the learning. After a lot of unsuccessful trials which got to the point of slight desperation, I finally struck gold. It turned out, the learning rate was too large so the loss function kept jumping around skipping the minima over and over again. I imagine something similar to this was happening to me:
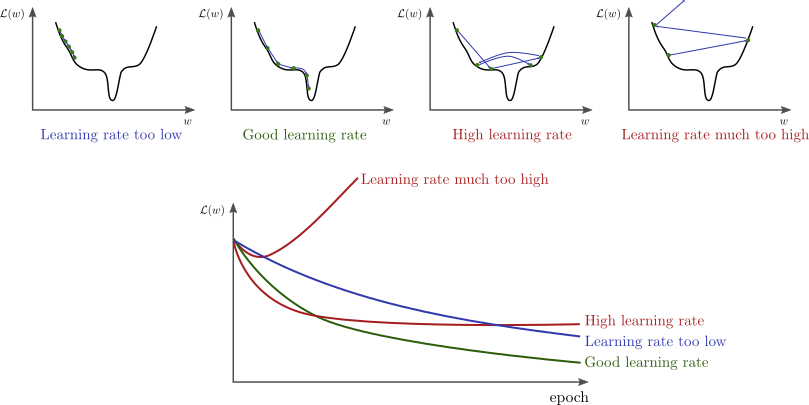
I want to note that I am using Adam optimizer so when I say learning rate I actually mean the initial learning rate, as Adam adjusts the learning rate for each weight based on an initial parameter. Anyways, once the learning rate was adjusted I started playing with the batch size, ending up with 16 as the optimal size.
However, up to this point overfitting kept being my #1 enemy. The model was able to deliver an almost perfect accuracy on the training set, but after a few epochs, this improvement wouldn’t be reflected on the validation partition.
I started researching how to reduce overfitting, and after trying a bunch of different ideas I found one that dramatically reduced it: introducing dropout layers between the dense layers of the classifier.
These dropout layers are filters that ignore a random fraction of the neurons during the training process. This adds noise to the training process and prevents the network from relying on very localized patterns and instead forces it to rely on different features every time, making the inference more robust.
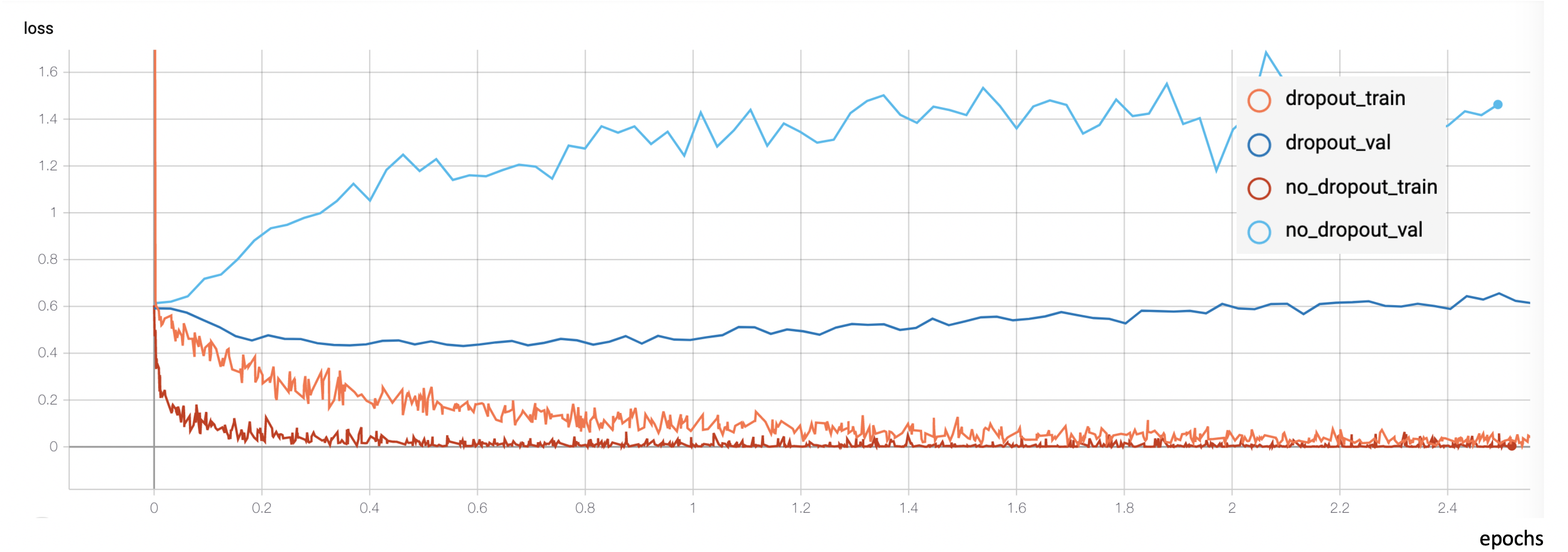
I ended up using a dropout layer before and after every dense one, each dropout layer dropping from 20 to 50 % of the input units. However, this wasn’t enough. The model was still overfitting after a few epochs. The last thing I tried which happened to also be very successful was further data preprocessing and augmentation. Let me explain. The original dataset I was working with consisted of 300K images. These images corresponded to frames of footage showcasing people undergo spontaneous emotional changes. However, due to the unbalanced nature of the dataset, I had to cap the most common classes so that every class would be equally represented. This reduced the dataset to about 200K images. But the network was still overfitting a lot, and what was especially weird about it was that the overfit happened before the model had seen all the images even once. A model cannot memorize data that hasn’t seen yet so something stinky was going on. After inspecting the images that the model was being fed I finally understood: contiguous frames in the videos were very similar to each other. So I was basically feeding the model the same image over and over again in each epoch. I ended up removing contiguous frames so only 1 frame out of every 5 was kept. This shrunk the dataset to 40K (quite a dramatic decrease, considering it originally was 300K).
This already gave me much better results but the last thing I tried was data augmentation: adding random modifications to each image every time they were fed to the model. These modifications consisted of random rotations, brightness shifts, shearings, zooms… Which in a way mimicked a larger dataset without repeated images.

To sum up, this process –which I really simplified for the sake of storytelling– led me to a model able to predict action units with a precision of .72 and a recall of .68 (evaluated on a balanced validation set). However, this is not the end of it. Emotions still need to be inferred from these action units. And this is not exactly a trivial process. So stick around for the next and final blog post next week explaining how I did this and how I deployed this pipeline to an application able to infer emotions in real-time!